All published articles of this journal are available on ScienceDirect.
Non-HLA Genes and Multiple Sclerosis
Abstract
Multiple sclerosis (MS) is an autoimmune demyelinating disease of the central nervous system. Identification of genetic variants that pose risks to MS is of high interest since they contribute largely to disease pathogenesis. A rich body of literature associated these risks with variants of HLA genes located mostly on the short arm of chromosome 6 (6p21). These genetic variations may result in alteration in protein function and are associated, therefore, with disease phenotype and therapy outcome. Although the HLA region has been routinely known to have the strongest correlation with MS, other genes found within and outside HLA locus are considered risk factors for MS. The objective of this review is to shed light on the non-HLA genes implicated with multiple sclerosis. Due to the interplay between the polygenetic and environmental factors, along with their differential contribution and genetic heterogeneity among populations, it is extremely challenging to determine the contribution of the non-HLA genes to the outcome and onset of MS disease. We conclude that a better assemblage of genetic factors involved in MS can have a critical impact on the establishment of a genetic map of MS that allows proper investigation at the expression and functional levels.
1. INTRODUCTION
Multiple sclerosis (MS) is an inflammatory neurodegenerative disorder of the central nervous system (CNS) resulting in a progressive loss of the myelin sheath that typically leads to sensory, motor, and neurocognitive disturbance [1]. According to the Atlas of MS, this unpredictable disease affects around 2.2 million people globally, under different clinical forms and manifestations [2]. Different data support the evidence that adults aged between 20 and 40 years are more susceptible to the disease, with a higher occurrence in females than males. Although MS etiology is poorly established, it is thought to be the consequence of environmental and genetic interactions [3]. Among environmental factors, smoking, infections, and sunlight exposure are the most widely reported [4-6]. Genetic factors related to MS are numerous and are believed to contribute to MS onset through various immunologically relevant genetic risk factors [7]. On one hand, HLA genes are believed to carry the strongest genetic risk variant for MS [8]. Consistently, it has been demonstrated that extended haplotypes spanning HLA class I and II are of high interest since they are implicated in the risk of several autoimmune diseases. On the other hand, non-HLA genes, including genes that may influence distinct immune tissues and cell types, contribute to the disease to a considerable extent. Recently, genome-wide associations studies performed by the International Multiple Sclerosis Genetics Consortium (IMSGC) analyzed genetic data from 47,429 MS patients and 68,374 individuals without MS in order to establish a genetic map for MS and have determined more than 230 independent MS risk variants, consisting of 200 autosomal variations, one chromosome X variation, and 32 different HLA ones. Various studies on different populations tackled the involvement of several non-HLA genes in MS with conflicting results. In this review, we will shed light on the mostly reported non-HLA genes along with their contribution to MS. These genes were chosen to be the major non-HLA contributors after a vigorous search conducted using Pubmed. In our search, we focused on related articles dated from January 2005 till January 2023.
2. NON-HLA GENES AND MS
Table 1 represents the main non-HLA genes that showed potential correlation with MS, along with their contribution to normal physiological conditions. These genes were the major non-HLA gene contributors obtained after a vigorous search conducted, using PubMed, from the published articles dated from January 2005 till January 2023.
| Non-HLA MS Susceptible Gene | Role in Normal Physiological Condition | References |
|---|---|---|
| CD58 | Co-stimulation and proliferation of T-cell receptor signaling via its interaction with cluster of differentiation 2 (CD2) | [9] |
| CD6 | Co-stimulation and proliferation of T-cell receptor signaling via its interaction with the ligand activated leukocyte cell adhesion molecule (ALCAM) | [10] |
| CLEC16A | Retrograde transport of HLA-II containing compartments | [11] |
| CYP27B1 | Implication in converting vitamin D to its active form | [12] |
| FoxP3 | Control T-cell activity by inducing the activity of regulatory T (Treg) cells | [13] |
| IL2-Rα | Binding of interleukin 2 (IL2) resulting in the activation of T cells | [14] |
| IL7-Rα | Binding of interleukin 7 (IL7) resulting in the differentiation of T cells | [15] |
2.1. Cluster of Differentiation 58 (CD58)
CD58 encodes for a glycosylated cell adhesion molecule known as lymphocyte-associated antigen 3(LFA3) that is found on human chromosome 1p13. It is present on the surface of antigen-presenting cells (APCs), especially macrophages, and is able to promote their specific adherence to the CD2 ligand on the T-cell surface. A whole-genome association scan has proposed that genetic variations in CD58 are associated with MS risk. The most studied genetic variant is rs2300747, where inconclusive data were shown; an association was maintained in different studies [16-18] and supported by the fact that MS patients carrying the G protective allele presented higher CD58 mRNA expression level during clinical remission [17]. However, this genetic variant association with MS was not consistent among other studies [19, 20]. On the other hand, a second genetic variant rs12044852 was shown to be correlated with MS in Australians and Russians [18, 21] but not in Malay and Iranian [19, 22] populations where the CC genotype was strongly associated with multiple sclerosis severity score (MSSS), indicating by this a negative effect of this SNP in response to interferon-beta (IFN-β) therapy [22].
2.2. Cluster of Differentiation 6 (CD6)
CD6 encodes a cell surface scavenger implicated in thymocyte differentiation as well as in T-cell activation and differentiation. It has been suggested that CD6 may play a crucial role in MS pathogenesis as it was shown to be involved in the transmigration of leukocytes across the blood-brain barrier (BBB). However, its definite role in regulating T-cell responses remains controversial. Genome-wide association studies have identified a large number of genetic variants associated with autoimmune diseases, including MS. Previous reports showed an association between CD6 genetic variant rs17824933 and MS [23-25]. In addition, the risk allele of this single nucleotide polymorphism (SNP) was demonstrated to be associated with a decreased expression of full-length CD6 in CD4+ and CD8+ T cells, affecting consequently their proliferation [26]. Hence, targeting CD6 by developing anti-human CD6 monoclonal antibody would be of high interest in MS therapy as it prevents T cell depletion [27]. Additional risk variants were shown to be associated with MS like rs12360861 in Poland population and rs650258 in the Spanish-Basque population [28, 29] with data replication needed. Overall, these data reinforce the important role of CD6 in MS pathogenesis.
2.3. C-type Lectin Domain Containing 16A (CLEC16A)
C-type lectins are key players in immune regulation as they drive different functions of antigen presenting cells (APCs) [30]. Located on chromosome 16p13, a susceptible locus for various autoimmune diseases, this gene is considered among the first non-HLA genes associated with MS [21, 31-33]. Moreover, upregulation of CLEC16A was observed in T cells of MS patients homozygous for the risk allele rs12927355 CLEC16A [34]. Additionally, higher expression of CLEC16A was detected in the white matter of MS patients, especially in the peripheral blood mononuclear cells (PBMCs). The high expression of this gene was blunted when patients were treated with vitamin D, indicating by this that CLEC16A may play a pivotal role in MS pathogenesis [11]. Additional functional studies are required to better understand the role of CLEC16A in MS as well as in other autoimmune diseases.
2.4. Cytochrome P450 Family 27 Superfamily B Peptide 1 (CYP27B1)
CYP27B1, located on chromosome 12q13-14, encodes for vitamin D metabolizing enzyme, the hydroxyvitamin D3-1-alpha-hydroxylase. Pre-vitamin D3 is produced in the skin and converted to 25(OH)D3 in the liver. In skin, kidney and immune cells, CYP27B1 enzyme converts 25(OH)D3 into 1,25 (OH)2D3 that binds to the vitamin D receptor present at the surface of T-cells and antigen presenting cells (APCs). Consequently, it suppresses the adaptive immune response, decreases dendritic cell and T-cell proliferation, differentiation, and maturation as well as Th1/Th2 ratio, and enhances the suppressive function of regulatory T-cells. Several studies have highlighted the role of rs703842 in MS with inconsistent results reported. Most studies showed an association between this risk variant and MS in Caucasian [35, 36], Slovakian [37], and Han Chinese [12] populations but not in others [38-40]. This was associated with a lower level of vitamin D registered in MS patients compared with controls, regardless of the risky genotypic and allelic status of this variant [35, 38]. One additional risk variant, rs118204009 was suggested to be implicated in MS [12, 41]. Further consideration of distinct ethnicity groups is needed for a better understanding of CYP27B1 role in MS.
2.5. Forkhead box P3 (FoxP3)
FoxP3 encodes a transcription factor that is predominantly expressed in CD4(+) CD25(+) regulatory T cells, playing a key role in maintaining immune homeostasis. It is considered the master transcription factor of these cells, responsible for the polarization of naïve T cells into Treg cells. Recently, T regulatory (Treg) cells have been known to present an impaired suppressive function in MS disease [42]. Accumulating evidence showed that functional alterations in FoxP3 gene expression have been observed in several autoimmune diseases [43, 44], linking thereby the defect in functional peripheral immunomodulation to an established genetic variant implicated in immune regulation and autoimmunity. A positive correlation was found between the genetic variant rs3761548 and MS [44-48]. Additional single nucleotide polymorphisms were also investigated with conflicting results [47, 50]. FoxP3 gene expression level was validated to be decreased in experimental autoimmune encephalomyelitis (EAE) models or in MS patients [51-53], which could not be reproduced in the study of Akbari et al. [54]. Interestingly, FoxP3 expression level could be restored by (IFN-β) treatment [55] or through injection of ex vivo autologous Treg cells that helped in increasing the level of Treg in patients’ blood. To sum up, FoxP3 could share a powerful link to MS susceptibility as it exerts an immunomodulatory effect.
2.6. Interleukin 2- receptor Alpha (IL2-Rα)
IL2-Rα, also known as CD25, is located on chromosome 10 and encodes the specific component of the high affinity IL2-R system that is implicated in autoimmunity and immune-regulation, where IL2/IL2-R signaling pathway allows the proliferation and survival of affected T cells and regulatory T cells production [56, 57]. Genome wide-association studies and fine mapping have revealed a tight link between SNPs in IL2-Rα and increased risk of immune mediated diseases including MS. Different studies were conducted on IL2-Rα polymorphisms and MS, mostly studying SNP rs2104286 that showed a risk susceptibility for the disease according to several publications [21, 58-61]. Additionally, one report has found that this SNP was accompanied by a reduced frequency of CD25(+) follicular helper T1 (TFH1) cells in patients carrying the risk genotype [62]. Fewer studies were performed on the rs12722489 SNP that also showed to be implicated with MS [59, 63]. However, data replication is needed on various populations for further validation. Furthermore, a study by Ainiding et al. showed that DNA hypo-methylation of IL2-Rα was significantly associated with MS and was accompanied by a higher expression of IL2-Rα in T cells of MS patients [64]. Together, these data confirm the tight correlation between IL2-Rα and MS.
2.7. Interleukin 7- receptor Alpha (IL7-Rα)
IL7-Rα, located on 5p13 human chromosome, encodes a subunit of IL7 receptor that plays a role in immune homeostasis by assisting in the maturation of B and T cells. Various genome wide association studies revealed that IL7-Rα is correlated with various immunological disorders [65], such as MS [66], and thus is considered among the top listed candidate genes implicated in MS. Evidence illustrates the tight association between MS and SNPs in the promoter and exon region of IL7-Rα. Genotyping of 123 SNPs in 66 genes chosen according to their chromosomal location or biological roles has identified that IL7-R includes at least 3 significantly associated SNPs with MS risk [67]. Additionally, alteration in the expression of genes encoding IL7-Rα and its IL7 ligand has been shown in the cerebrospinal fluid (CSF) compartment of MS patients [68]. Most studies tackled the association between genetic variant located in exon 6 of IL7-Rα and MS with a well-established association [63, 69-77] or association reach-ing significance after stratification analysis in progressive MS subjects only [78, 79] or more specifically in secondary- progressive MS (SPMS) [80]. However, no association was found in other studies [81-83]. It has been suggested that rs6897932 risk variant is linked to altered alternative splicing of exon 6 that contributes to its skipping, affecting therefore, the ratios of soluble (sIL7- Rα) to membrane-bound IL7-Rα [84]. However, a reduced expression of sIL7-Rα was detected in progressive MS subjects regardless of their genotypes [78] and showed no effect of genotype or protein isoforms expression on MS phenotype [71]. Furthermore, a genetic variant in the 5’UTR of the RNA helicase DDX39B, a potent activator of exon 6 of IL7-Rα, was shown to reduce its translation, resulting in increased levels of the soluble form of IL7-Rα [85-88]. A meta-analysis study consisting of 9734 cases and 10436 controls confirmed the association between 3 SNPs of IL7-Rα (rs3194051 in exon 8, rs987107 in the intronic region, and rs11567686 in the promoter) and MS [66]. To sum up, IL7-Rα locus polymorphisms can have a key role in MS predisposition.
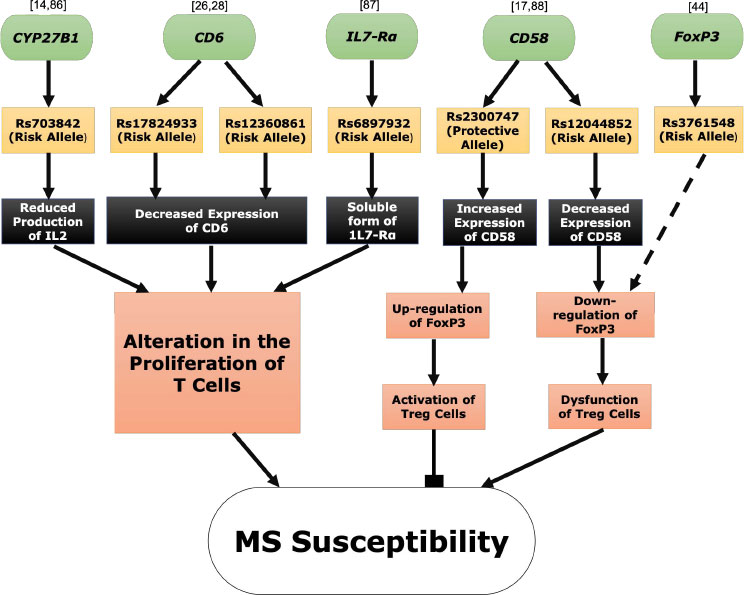
Fig. (1) depicts a functional analysis of the main non-HLA MS susceptible genes and their potential contribution to MS.
CONCLUSION
Multiple sclerosis is a chronic autoimmune disorder where genetic variations, especially those involved with immune regulation, play a key role in its development. The literature on HLA related genes in MS is rich, including genetic and functional studies. Importantly, the latter showed that HLA genetic associations with MS were linked to functions not directly associated with antigen presentation. On the other hand, although non-HLA genetic variations were prominently associated with MS, conflicting results were reported based on the genetic backgrounds. Thus, more studies are needed to verify the functional impact of these variants in MS progression. In this review, we summarized the most relevant non-HLA genetic variants associated with MS. Overall, these variants may serve as a starting point for MS genetic map construction for a further investigation of these genes at transcriptional, functional, and signaling pathway levels.
LIST OF ABBREVIATIONS
| MS | = Multiple Sclerosis |
| CNS | = Central Nervous System |
| IMSGC | = International Multiple Sclerosis Genetics Consortium |
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


