All published articles of this journal are available on ScienceDirect.
Transferability of Sorghum Microsatellite Markers to Bamboo and Detection of Polymorphic Markers
Abstract
The use of molecular markers for the characterization and evaluation of plant genetic resources has become a useful approach in plant genetic research. Simple Sequence Repeats (SSRs) are among the markers that are widely used in genetic diversity and parental analysis owing to their co-dominant nature, high reproducibility, abundance in the genome and transferability across species or genera. The development of these markers for a species might be costly and time consuming. Hence, screening existing markers through transferability test from closely related species or family is resource conscious. In this study, the transferability of 90 polymorphic SSR markers of sorghum to bamboo was tested and polymorphic analysis of transferable markers were performed. Nearly 62% of the tested SSRs successfully recorded amplification in at least one bamboo species of which 55% were polymorphic. These polymorphic markers detected a total of 147 alleles at an average rate of 4.7 alleles per marker. The abundant alleles account 20.4% while the common and rare alleles share 39.6 and 40 %, respectively. The result showed a relatively low degree of polymorphic information content (PIC) averaging 0.29. The gene diversity index (He) ranged from 0.21 to 0.49 with a mean of 0.37. The cluster analysis based on the polymorphic markers surfaced most of the species in accordance with their geographic origin. The complementarity of the weighted neighbour joining tree and coordinate analysis implies the representative nature of the transferred markers for the diversity analysis of bamboo species.
INTRODUCTION
Bamboos are members of the sub-family Bambusoideae within the grass family Poaceae [1] in which most important cereal crops such as rice, wheat, sorghum, maize and barley are also grouped. Bambusoideae is a large sub-family containing more than 70 genera with over 1450 species [2]. It is a fast growing wood grass distributed all over the world, but major contributors of bamboo resource are Asia (particularly China, India Japan, Myanmar and Malaysia) and South America mainly Brazil, Venezuela and Colombia [3]. Ethiopia ranks first in Africa which comprises 67 % of the total bamboo area coverage of the continent [4].
Bamboo is an important multipurpose genetic resource in the world for its wide range of economic values. It is the source of building materials, high quality furniture, food, fiber, fodder, pulp, paper, board and charcoal [5-8]. It also serves as ornamental plant. The plant also plays a significant role in ecological applications because of unlimited environmental values [2]. Due to its fast growth nature, it can be harvested within short period of time without depletion and deterioration of the soil. It can grow on marginal land which is not suitable for agriculture or forestry. In addition, the plant serves as source of feed for many wild animals.
Despite its huge importance of the plant, it is less studied and its genetic diversity is little understood. Besides, very wide area coverage in western and south western Ethiopia currently are severely affected as a result of mass flowering and seed setting of the plant [9]. It demands extensive research to exploit the effective methods for characterization and conservation of this important genetic resource. Morphological traits are the most commonly used approach in bamboo classification [3]. Although morphological traits remain to be major characterization tools, the recent advance in molecular genetics has enabled in refining classification and within species diversity analysis at higher resolution.
Various marker systems such as random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), diversity arrays technology (DArT) and single nucleotide polymorphism (SNP) have been used in the study of various plant genetic resources. Owing to the technical simplicity in their application, recently, SSRs have become major tools in population genetics, genetic mapping and other plant genomic studies [10]. These markers are highly reproducible, which is highly important in genetic analysis. They are highly polymorphic and produce very high allelic variations even among very closely related varieties. The co-dominance nature of the marker also helps to analyze the segregating populations or paternity testing. The SSR markers are abundant and well distributed throughout the nuclear genome of the species [11, 12]. However, the development of SSR markers requires relatively high cost and time consuming [13, 14]. On account of their transferability across related families, SSR developed for one species can be used for other species within same family.
Microsatellites are usually transferable across grass family due to the evolutionary relationships among species belong to the same family. SSR markers particularly expressed sequence tag (EST)-derived SSRs have shown to possess a higher potential for inter-specific transferability than genomic SSRs [15]. Several studies have been conducted on the transferability of SSR markers across species or genera of several cereal crops which belong to grass family such as rice [16], wheat [17], wheat, rye and triticale [18], barley [19], sugar cane [20, 21], sorghum [22, 23] major cereal to minor grass [24] and pearl millet [25]. There are also reports about transferability of SSRs from cereals such as rice and sugar cane to bamboo species [26, 27]. Sharma et al. [15] also conducted an experiment on identification and amplification of EST-SSR markers in different bamboo species. Sorghum is one of the cereals crops with already developed large number of SSRs markers and it is genetically closer to bamboos than other cereals such as rice. However, there has been no report on transferability of sorghum SSR markers to bamboo species. The objective of this study was to perform the transferability test of SSR markers from sorghum to bamboo species and to employ the transferable markers in the genetic diversity analysis of selected bamboo species.
MATERIALS AND METHODS
Plant Materials and DNA Isolation
Fresh leaf tissues from seven exotic and two indigenous Ethiopian bamboos representing both temperate and tropical species were collected (Table 1). Five leaves from each of the species were bulked and lyophilized prior to DNA extraction. Genomic DNA was extracted using Wizard® Genomic DNA Purification Kit (Promega, USA). Genomic DNA samples from sorghum were also included as control. The DNA concentration was quantified by spectrophotometer (Thermo Scientific, USA) and the quality was checked using 0.8% (w/v) agarose gel stained with GelRed® (Biotium, USA). The samples were dissolved with pure sterilized water and the final volume was diluted to 10 ng/µl. Both genomic and PCR product bands were viewed using gel image system (Uvitec, UK).
Bamboo species that are used as test genotypes.
| Species | Morphological features | Source | Origin/ Native to |
|---|---|---|---|
| Gigantochloa apus | Strongly tufted woody bamboo with erect drooping culms that can reach a height of 8-22 m. The culms are bright green or yellowish-green when young, with an average diameter between 4-13 cm and a wall thickness between 6-13 mm. Many clustered branches at the nodes with one larger dominant branch. | HARC | Malaysia |
| Gigantochloa atter | Large tufted woody bamboo with dark green culms of 15-22 m high and 5-10 cm in diameter. Culm internodes are thin-walled and are on average 40-50 cm long. Many clustered branches at the nodes with one larger dominant branch. | HARC | Indonesia |
| Phyllostachys bambusoids | It can reach a height of 15-22 m and a diameter of 10-15 cm. The culms are dark green, quite thick and very straight. Leaves are dark green. The flowering interval of this species is very long, reaching about 130 years. | HARC | China/Asia |
| Guadua angostifolia | It is characterized by a high growth rate (11 to 21 cm per day). It is an ideal construction material with a high percentage of fibers, a specific gravity of 0.5 to 0.6 and excellent structural properties such as a high resistance to-weight-ratio, a high capacity to absorb energy and excellent flexibility | HARC | South America |
| Guadua amplexifolia | Characterized by a solid culm in the base and a small lumen in the distal part. | HARC | South America |
| Bambusa textiles | Very thin culms and tight clumps. Arches are very gracefully. | HARC | China |
| Gigantochloa sumatra | Characterized by the edible young shoots | HARC | Asia/Sumatra |
| Arundinaria alpina | The internodes are smooth and cylindrical, but slightly tapering and sheaths are usually not shed. | HARC | Africa |
| Oxytenanthera abyssinica | Clump-forming bamboo with a robust rhizome up to 10 cm in diameter with dense clums typically consisting of 20-100 culms. The plant can reach a height of 5-15 m and a diameter of 3-10 cm. The internodes the length of the internodes is between 15-40 cm. | HARC | Africa/Ethiopia |
HARC (Holetta Agricultural Research Center) is one of the oldest and biggest Agricultural Research Centers in Ethiopia located 38°30′E, 9°4′N with an altitude of 2400 m.a.s.l. receiving annual average rainfall of 1100 mm with minimum and maximum temperature of 6 & 22 ºC, respectively.
Selection of Sorghum SSR Markers and PCR Analysis
A pair of 90 SSR polymorphic sorghum primers [28-31] that are evenly distributed across the whole sorghum nuclear genome were selected to test the transferability of the marker across the selected bamboo species (Table 2). PCR amplification was performed in a 10 µl reaction volume comprising of 1 x PCR buffer (20 mM Tris-HCl, pH 7.6; 100 mM KCl; 0.1 mM EDTA; 1 mM DTT; 0.5% (w/v) Triton X-100; 50% (v/v) glycerol), 2 mM MgCl2, 0.16 mM dNTPs, 0.16 µM fluorescent labeled M13-forward primer, 0.04 µM forward primer, 0.2 µM reverse primer, 0.2 units of Taq DNA polymerase (SibEnzyme Ltd, Russia) and 30 ng of template DNA. All forward primers contained an M13-tag (5’- CACGACGTTGTAAAACGAC - 3’) on the 5’ end that was fluorescently labeled to allow detection of amplification products. The Forward primers were labelled with FAM, PET, NED or VIC (Applied Biosystems). PCR reactions were carried out in a GeneAmp® PCR System 9700 thermal cycler (Applied Biosystems, USA) programmed for initial denaturation at 94°C for 15 min, followed by second denaturation at 94°C for 30 sec, annealing temperature ranging from 50°C to 55°C for 1 min, extension at 72°C for 2 min and final elongation at 72°C for 20 min.
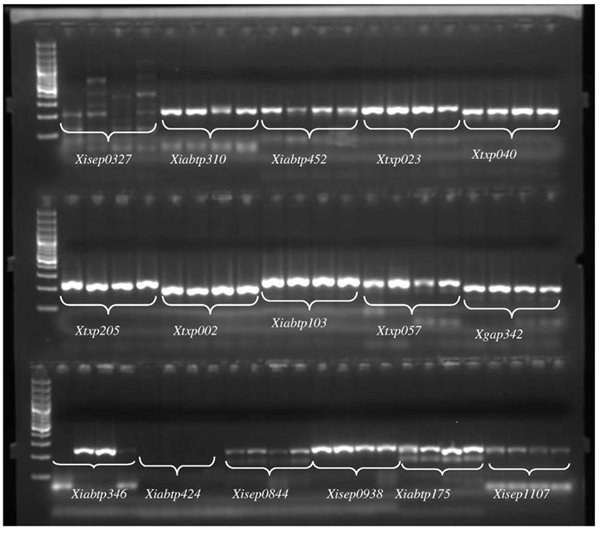
Informative sorghum microsatellites showing high rate of transferability across the tested bamboo species. Amplicons were generated from genomic DNA of Arundinaria alpine by PCR at 55 °C annealing temperature. The fragment was separated on 2% (w/v) agarose gel along with 1 kb ladder. Xiabtp424 didn’t show any band and hence used as a negative control.
Successful amplification was confirmed by running 2.0 µl of the PCR products on a 2% (w/v) agarose gel stained with GelRed® (Biotium) and visualized using gel image system (UVtech, UK). Annealing temperature for the selected bamboo species were optimized in order to obtain clear bands.
Clear and strong bands were observed at annealing temperature 55°C (Fig. 1) while the optimum annealing temperature for most sorghum polymerase chain reaction is 50°C. The success of PCR amplification is largely dependent on the annealing temperature of the PCR program. Optimization of the program for the selected experiment is a crucial step during the analysis. The optimized annealing temperature (55°C) can be applied to amplify the DNA of various bamboo species and other related polyploidy species.
| No. | Marker Name |
Forward Primer | Reverse Primer |
|---|---|---|---|
| 1 | gpsb151 | ATACCAAGTTTCCTTTACCT | GTTGGGGGAGAGTTTT |
| 2 | SbAGE01 | GACCGATCTAATGATGCAG | ACGGTAGAGAAAGACCCATC |
| 3 | SbAGH04 | GGCACTCATGGAGTCACA | TTTATCCAAATCAAACCGG |
| 4 | SbKAKG1 | AGCATCTTACAACAACCAAT | CTAGTGCACTGAGTGATGAC |
| 5 | Xcup02 | GACGCAGCTTTGCTCCTATC | GTCCAACCAACCCACGTATC |
| 6 | Xcup16 | TGCAGTGCTAGCTCATGGTC | CTTTCCAGCCTCCCATATCC |
| 7 | Xcup24 | AAACTGGATGCCACACCAAG | AGCTATACCAACACGGGCAG |
| 8 | Xcup33 | GCGCTGCTGTGTGTTGTTC | ACGGGGATTAGCCTTTTAGG |
| 9 | Xcup36 | TGAGCTGATAATGGCTGCTG | GCGTCACGGAAGTTGGAC |
| 10 | Xcup40 | ACGGAGAATAGAAAGTGGCG | TTGAGCATGCAACCACCTAC |
| 11 | Xcup50 | TGATTGATTGAGGCAGGCAC | TTCCGGTCTCTGTCCATTTC |
| 12 | Xgap001 | TCCTGTTTGACAAGCGCTTATA | AAACATCATACGAGCTCATCAATG |
| 13 | Xgap084 | CGCTCTCGGGATGAATGA | TAACGGACCACTAACAAATGATT |
| 14 | Xgap342 | TGCTTGTGAGAGTGCCTCCCT | GTGAACCTGCTGCTTTAGTCGATG |
| 15 | Xisep0101 | CAGATCTCCGGTTGAAGAGC | TGAGCCGAGCTCAACATACA |
| 16 | Xisep0108 | GTACGTTCCCCATCCTTCCT | CTCCTGTTCTCTCCGCATTC |
| 17 | Xisep0110 | GAGGGGAAGCTGGAGACC | TCAAGTGTACAACGCATCCAG |
| 18 | Xisep0138 | GAGATCGAGAGGCACTTTGG | CAGCGACAAGCCAATACCA |
| 19 | Xisep0224 | ACTGGGGTTCCTTTTCCTGT | TCCCTGATTTCCCCTCTTTT |
| 20 | Xisep0327 | CTGTTTGTGCTTGCAACTCC | TCATCGATGCAGAACTCACC |
| 21 | Xisep0422 | TGCCCGTAATTAAGCCCATA | CCCACTGCTCCAGGTAAGAA |
| 22 | Xisep0429 | GTCGTCTGGAAGCAACAGC | TGGGGGTAGTTGGTGGTG |
| 23 | Xisep0443 | TCATGTACAGAGGCGACACG | AGGTCGCAACAGACACCTTC |
| 24 | Xisep0449 | CCGCTCATCAGTCATCACAT | ACAAAATCCATCCCACAACG |
| 25 | Xisep0511 | CCTCGCCCAAAACCCTAC | GAGGATCACCTCATCGTGCT |
| 26 | Xisep0539 | GACCCCATCTCTTCCTTTCC | AGACTGAGGCACCGCTTG |
| 27 | Xisep0543 | CGAGGGTTTTTCTTCTGTGG | GACATCGGAGACCTTGAGGA |
| 28 | Xisep0612 | CTCTCTGTCCTCCTCGTCGT | CCTGCTTCTTGGACACCTTC |
| 29 | Xisep0617 | GGCTGGGAGAGCTAGGAAGA | GACGGCTCGTCCATCATC |
| 30 | Xisep0622 | GAGGATCGGAGGAAGAGACC | TCTCCCATTCTCCCCTCTTT |
| 31 | Xisep0639 | TCGGACGGAGTCATCAGATA | GCCTTCGTGTCTTCTGTCCT |
| 32 | Xisep0648 | GAGAAGTTGGAGCGGAGGA | AACACCCAGATCAGCGAAAC |
| 33 | Xisep0805 | CTCCCCCGTGATTTGATCT | TAAGCAAAAGCACCATCAGC |
| 34 | Xisep0831 | TCCATGACCTTGAGGAGGAG | TTGAAGCAGGACAACACACC |
| 35 | Xisep0839 | TACGCATAGCGCCTTTCAAT | ATTTCATTATGCCGGTCTCG |
| 36 | Xisep0938 | TGCTGTTCTTGAACGTGTTTG | TTTTGCACAAAGTTGCGTGT |
| 37 | Xisep1029 | GACCCTCCTCCTCAACCACT | CATGCATGCACAAGCAGATT |
| 38 | Xisep1032 | GCAAGCTCTACGGGATCTTC | GCAGCTGGAAAATAATCGAAA |
| 39 | Xisep1039 | GTGGATTCAAATCCGCTGAC | GGCAATTTGGCAAGCAAT |
| 40 | Xisep1042 | GGAGGCAAGTTCAGGAAGTG | TGTGTGCAGTGCATGCTTAG |
| 41 | Xisep1107 | GGATAATCTGCAGGCGACTT | CCATCTGCTGCTCTGACTTG |
| 42 | Xisep1208 | TCCAAACACACAGACCGTTT | TCCGATGGTTGAGAGCTTGT |
| 43 | Xisep1231 | CTGCTTATGCGCTTCGATTT | CATAATGGGTGCACTCTAGCC |
| 44 | Xtxp008 | ATATGGAAGGAAGAAGCCGG | AACACAACATGCACGCATG |
| 45 | Xtxp012 | AGATCTGGCGGCAACG | AGTCACCCATCGATCATC |
| 46 | Xtxp014 | GTAATAGTCATGACCGAGG | TAATAGACGAGTGAAAGCCC |
| 47 | Xtxp023 | AATCAACAAGAGCGGGAAAG | TTGAGATTCGCTCCACTCC |
| 48 | Xtxp024 | TTGTGTAGTCCATCCGATGC | TTCTAAGCCCACCGAAGTTG |
| 49 | Xtxp026 | AAGTGTAGTAGCAGTTTAGTCTC | TAGGTATCAAAGGACCAAGG |
| 50 | Xtxp030 | AAAAAGGACGCGCAGCTG | CTGGTCTCCACCATCCGTAG |
| 51 | Xtxp033 | GAGCTACACAGGGTTCAAC | CCTAGCTATTCCTTGGTTG |
| 52 | Xtxp034 | TGGTTCGTATCCTTCTCTACAG | CATATACCTCCTCGTCGCTC |
| 53 | Xtxp040 | CAGCAACTTGCACTTGTC | GGGAGCAATTTGGCACTAG |
| 54 | Xtxp057 | GGAACTTTTGACGGGTAGTGC | CGATCGTGATGTCCCAATC |
| 55 | Xtxp088 | CGTGAATCAGCGAGTGTTGG | TGCGTAATGTTCCTGCTC |
| 56 | Xtxp094 | TTTCACAGTCTGCTCTCTG | AGGAGAGTTGTTCGTTA |
| 57 | Xtxp113 | CTCAGCTAATTTAGCCATAG | CAAGTAATAGACGAGTGAAAG |
| 58 | Xtxp141 | TGTATGGCCTAGCTTATCT | CAACAAGCCAACCTAAA |
| 59 | Xtxp159 | ACCCAAAGCCCAAATCAG | GGGGGAGAAACGGTGAG |
| 60 | Xtxp205 | CCTGCCGTGTCTTCC | TATATGCATGCCGTAGATTT |
| 61 | Xtxp211 | TCAACGGCCAATGATTTCTAAC | AGGTTGCGAATAAAAGGTAATGTG |
| 62 | Xtxp258 | CACCAAGTGTCGCGAACTGAA | GCTTAGTGTGAGCGCTGACCAG |
| 63 | Xtxp270 | AGCAAGAAGAAGGCAAGAAGAAGG | GCGAAATTATTTTGAAATGGAGTTGA |
| 64 | Xtxp274 | GAAATTACAATGCTACCCCTAAAAGT | ACTCTACTCCTTCCGTCCACAT |
| 65 | Xtxp279 | ATTCTGACTTAACCCACCCCTAAA | AGCTCATCAATGTCCCAAACC |
| 66 | Xtxp283 | CGCCCGAACTCTTCTTAAATCT | ATTATGCCCTAACTGCCTTTGA |
| 67 | Xtxp284 | CCAGATTGGCTGATGCATACACACT | AAGGGTAATTTATGCACTCCAAGGTAGGAC |
| 68 | Xtxp285 | ATTTGATTCTTCTTGCTTTGCCTTGT | TTGTCATTTCCCCCTTCTTTCTTTT |
| 69 | Xtxp286 | AGCAGCAGCAGCAACAG | GCGTGGTCTTTGTGGTTC |
| 70 | Xtxp295 | AAATCATGCATCCATGTTCGTCTTC | CTCCCGCTACAAGAGTACATTCATAGCTTA |
| 71 | Xtxp312 | CAGGAAAATACGATCCGTGCCAAGT | GTGAACTATTCGGAAGAAGTTTGGAGGAAA |
| 72 | Xtxp320 | TAAACTAGACCATATACTGCCATGATAA | GTGCAAATAAGGGCTAGAGTGTT |
| 73 | Xtxp348 | CGACATCAGCGTTGTCTTTCTA | GCTTACGAATAGGGCAAAAGAACT |
| 74 | Xtxp355 | TGGTTGGAAAGATAATCAAG | GCCCTAATGAGTCCTCAC |
| 75 | Xtxp357 | CGCAGAAATACGATTG | GCTATCTGGAGTAACTGTGT |
| 76 | Xiabtp 346 | CCGTCTCCACAAGCTTCTTC | GACTGTGCCAGCTGTCTTCC |
| 77 | Xiabtp 452 | CATGGATCGACCCTTTTGTTC | AGCGACCGAGCACAACTTATC |
| 78 | Xiabtp 175 | ATGCACCAGCTCAAGGCTATC | ACGTGCATACGTGCAGAGTC |
| 79 | Xiabtp 361 | TTTGCTGGACACTGAAAACG | ATCATGCATCCATGTTCGTC |
| 80 | Xiabtp 310 | CACAAGACACGCACAAAGGTG | GAGAAGCATTTGCCATGGATG |
| 81 | Xiabtp 240 | GAGCACAGATTTCTTCCTCCTC | TTATCTGCGACAAACCCACA |
| 82 | Xiabtp 26 | CGCACTCGATCGACTAGCTTA | CGAGAGAGGCGATAAGGATC |
| 83 | Xiabtp 106 | CACCCTCCTCTCATCCTCAC | GCGCCGTAGTCGTAAAGG |
| 84 | Xiabtp 424 | ACCTGCAGCCTACCACAGATC | AATCCTTTTTCGTTGCCAGA |
| 85 | Xiabtp 247 | GTCCAAGGCTTTCAATCTGC | TGGGACAGCAGATTACGACA |
| 86 | Xiabtp 340 | CATTGCTCACTGCTCAGTTCA | CCATCGATCGAGCTCTCTG |
| 87 | Xiabtp 103 | CACTTCGTTGTGCCACATTC | ACTCTGCCACCCAATCAAAC |
| 88 | Xiabtp 168 | AAGAAGGACAAGAAGCGGAAG | GCTCTTCACGGTCTCCTCTG |
| 89 | Xiabtp 378 | GAGCAGOGGATAAGTTCGAG | TAACCACATGGCCCGTTC |
| 90 | Xiabtp 69 | TGCACAAGAGGCAGGATATG | AGCCCATGAGAAAAAGCTCA |
In order to perform the genetic diversity and relatedness among the target species, the successfully transferable markers were genotyped using capillary electrophoresis. Depending on the nature of fluorescent label and strength of the amplification bands used, a volume ranging from 2.5 µl to 3.5 µl of four different amplification products were co-loaded along with the internal size standard, GeneScan™ -500 LIZ® (Applied Biosystems) and Hi-Di™ Formamide (Applied Biosystems, USA). The fragments were denatured at 95°C for five minutes using GeneAmp® PCR System 9700 thermal cycler. The fragments were separated by capillary electrophoresis using an ABI Prism® 3730 Genetic analyzer (Applied Biosystems) in order to determine the exact allele size of each fragment.
Data Analysis
PCR fragment size was manually scored using GeneMapper v4.0 software. The allele size in terms of base pairs was compared to the internal size standard, GeneScan™ -500 LIZ® (Applied Biosystems, USA). Percentage of transferability was computed by dividing the number of the SSR markers amplified in non-donor (bamboo) species to the total number of tested SSRs. Fragments in base pair were converted to binary data because of the complex amplification pattern in bamboo genome [27]. The level of polymorphism of the entire markers across the selected bamboo species was computed. Only polymorphic markers were selected to analyze the genetic diversity and relatedness among the target species. The proportions of polymorphic loci were calculated as:
Where,
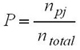 |
P = proportion of polymorphic loci
npj = number of polymorphic loci
ntotal = total number of loci
Power Marker v.3.25 [32] was used to compute PIC and allele frequencies. Rare, common and abundant were manually computed in MS Excel (Microsoft Inc., Seattle, USA). Polymorphism information content (PIC) was calculated using the method of Botstein et al. [33].
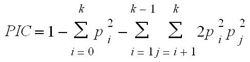 |
Where, pi and pj are the frequencies of alleles i and j, respectively
Markers with a PIC value of more than 0.5 were considered highly informative, between 0.25 and 0.5 as informative and less than 0.25 as less informative.
Pair-wise genetic differentiation between the selected bamboo species, a genetic dissimilarity matrix was calculated using simple matching with DARwin v5 software (available at http://darwin.cirad.fr/darwin/Home.php). Individual relations were analyzed with a tree construction based on Neighbor Joining (NJ) method, as implemented in DARwin v5.
RESULTS AND DISCUSSION
The availability of large number of sorghum SSR markers as a community resource and the consideration of sorghum as a model crop to study other grass family created a scope of opportunity to exercise similar methodology in non-model plants like bamboo. The current advancement in genomic technology will also help to set the best conservation strategies particularly for the endangered plant genetic resource as well as economically important plants like bamboo. To achieve this magnificent plan of genetic resource conservation, it is imperative to clearly characterize the core collection prior to conservation practices.
Among 90 tested sorghum SSR markers, 62.2% of them scored successful amplification in a bamboo representative species, Arundinaria alpine (Table 3). However, transferability rate differed among bamboo species. The lowest transferability rate that accounted only 40% was observed in Gigantochloa atter. Markers with high rate of transferability include Xtxp023, Xtxp205, Xtxp283 and Xcup16 where as Xiabtp346 and Xisep0327 showed reduced transferability rate. Markers such as Xiabtp424, Xise0443 and Xisep0648 did not score any amplification in either of the bamboo species. The transferability rate obtained in this study is higher than the one reported for various grass family species including bamboo using [18, 26, 27]. However, the rate is slightly lower than the rice SSR transferability to bamboo that was reported to be 68% [26]. It is possible to improve the transferability rate of markers by using markers that were developed from expressed sequences [18].
The scored fragment size ranged between 104 bp and 486 bp. Most of these fragments were amplified higher than the size of the donor fragment, which is in agreement with the previous study implying that the amplified fragments from the selected species have different allele sizes than those of the donor species [26, 24, 27].
Out of the 56 SSR markers that were found to be transferable, 31 (55%) of the showed polymorphism. A total of 147 SSR marker alleles were recovered, at an average rate of 4.5 alleles per marker. The average number of fragments per marker is less than the previous transferability test conducted using rice and sugar cane SSRs markers to study bamboo genetic diversity [27]. The number of alleles per marker ranged from 2 to 12 with the highest number of alleles obtained from Xisep0138 followed by Xcup24 and Xiabtp240 (Table 4). The presence of these multiple amplification products could be due to duplication, high level of polyploidy and genetic variability in bamboo species [12, 27, 34]. Among the polymorphic markers (n=31), 15 of them showed a clear band and highly transferable to all the bamboo species tested in this experiment (Fig. 1). These markers are found to be very effective and useful resources to utilize them for phylogenetic and genetic diversity studies of the different bamboo species. In contrary, very few markers were amplified only in some specific bamboo species ( Supplementary Table S1).
A total of 59 alleles (40 %) were detected as rare alleles with an average of two alleles per marker. The highest number of rare alleles was obtained for marker Xcup24. Similarly, 58 alleles (39.5%) scored common allele frequency considered as common alleles with an average of two alleles per marker. Xisep0327 showed the highest number of common allele. The remaining 30 alleles (20.5%) were found to be abundant allele with an average number of one allele per marker. The higher frequencies of rare and common alleles are very important resources for future bamboo characterization and maintenance program. Even though, the significance of the rare alleles cannot be speculated from the current results, their abundance in the selected bamboo species suggests that this diverse set of genetic resource has not been extensively exploited.
The level of polymorphism detected across species or genera mainly depends on the genetic divergence of species tested and primers used [24]. The PIC values for the SSR loci ranged from 0.19 to 0.37 with an average of 0.29 (Table 4). Five markers such as Xtxp205, Xtxp057, Xiabtp310, Xtxp040 and Xiabtp340 showed the highest PIC value of 0.37 whereas the lowest PIC value of 0.19 was recorded for marker Xcup24. The gene diversity index (expected heterozygosity, He) ranged from 0.21 to 0.49 with a mean of 0.37. Marker Xcup24 presented the lowest gene diversity as well as PIC value. Though the computed average PIC value in this experiment was relatively smaller, it would be sufficient to utilize in characterization of bamboo collection. The wide variation in ploidy level among the different bamboo species has created the generation of multiple alleles derived from a single locus. This has implication that a small number of markers can discriminate the different bamboo species and a useful aspect towards the conservation of the genetic resources with rare allele.
Rate of sorghum SSR transferability across the selected Bamboo species.
| Species Name | Number of amplified markers | Percentage of amplification |
|---|---|---|
| Gigantochloa apus | 46 | 51.1 |
| Gigantochloa atter | 36 | 40.0 |
| Phyllostachys bambusoids | 50 | 55.6 |
| Guadua angostifolia | 42 | 46.7 |
| Guadua amplexita | 48 | 53.3 |
| Bambusa textiles | 54 | 60.0 |
| Gigantochloa sumatra | 46 | 51.1 |
| Arundinaria alpina | 56 | 62.2 |
| Oxytenanthera abyssinica | 48 | 53.3 |
The highest genetic distance (0.90) was observed between Guadua amplexita, Gigantochloa apus and Guadua amplexita, Gigantochloa atter while lowest genetic distance (0.32) was recorded between Gigantochloa apus and Gigantochloa sumatra (Table 5). Relatively higher genetic distance (0.77) was obtained between Ethiopian lowland (Oxytenanthera abyssinica) and highland bamboo (Arundinaria alpine). The distinctiveness of these species requires proper conservation techniques in order to maintain the genetic resources.
Genetic analysis based on the polymorphic markers successfully grouped the different species in accordance to the geographic origin of the species and their genera (Fig. 2). This study revealed that Ethiopian bamboos which exist in different genera are highly differentiated. The dandrogram generated from Neighbor-Joining (NJ) tree analysis was in congruent with the result obtained from the principle coordinate analysis with few exceptions (Figs. 2, 3). The similarity in the clustering of these bamboo species using these two approaches further validates the representative nature of the transferred markers for the diversity analysis of bamboo species. The current study will serve as a foundation to study bamboo genetic resources using the transferable markers and hence exhaustive collection and characterization of both highland and lowland bamboo from the entire growing regions will be necessary in the future.
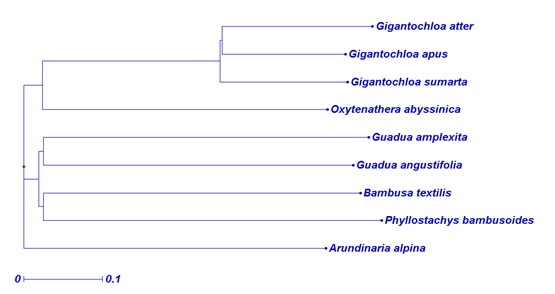
Phylogenetic groups of selected bamboo species based on selected 31 polymorphic SSR loci.
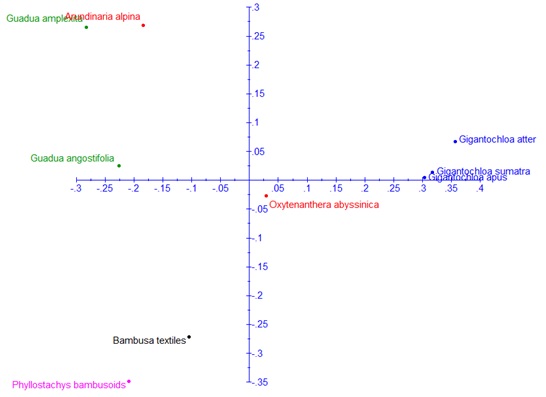
Biplot of the axis 1 and 2 of the principle coordinate analysis based on the dissimilarity of 31 SSR markers.
Basic statistics of 31 sorghum microsatellites markers that are found to be polymorphic and transferable to selected bamboo species.
| Marker | Total alleles | Rare alleles | Common alleles | Abundant alleles | Gene diversity | PIC |
|---|---|---|---|---|---|---|
| Xisep0824 | 5 | 2 | 2 | 1 | 0.31 | 0.25 |
| Xisep0327 | 8 | 1 | 7 | 0 | 0.36 | 0.29 |
| Xisep0138 | 12 | 9 | 3 | 0 | 0.26 | 0.22 |
| Xtxp205 | 3 | 0 | 3 | 0 | 0.49 | 0.37 |
| Xtxp088 | 3 | 1 | 2 | 0 | 0.30 | 0.25 |
| Xcup16 | 3 | 1 | 0 | 2 | 0.30 | 0.25 |
| Xtxp094 | 7 | 3 | 3 | 1 | 0.30 | 0.25 |
| Xtxp030 | 6 | 4 | 2 | 0 | 0.29 | 0.24 |
| Xisep0844 | 2 | 0 | 1 | 1 | 0.44 | 0.35 |
| Xtxp320 | 2 | 0 | 1 | 1 | 0.42 | 0.33 |
| Xtxp295 | 5 | 1 | 4 | 0 | 0.34 | 0.28 |
| Xtxp057 | 2 | 0 | 1 | 1 | 0.49 | 0.37 |
| Xcup24 | 11 | 10 | 1 | 0 | 0.21 | 0.19 |
| Xisep0938 | 5 | 1 | 0 | 4 | 0.41 | 0.32 |
| Xiabtp452 | 5 | 0 | 2 | 3 | 0.41 | 0.32 |
| Xiabtp361 | 2 | 0 | 0 | 2 | 0.35 | 0.27 |
| Xiabtp310 | 2 | 0 | 1 | 1 | 0.49 | 0.37 |
| Xisep1107 | 3 | 1 | 0 | 2 | 0.38 | 0.30 |
| Xiabtp240 | 9 | 6 | 3 | 0 | 0.29 | 0.24 |
| Xtxp040 | 2 | 0 | 0 | 2 | 0.49 | 0.37 |
| Xtxp002 | 3 | 1 | 1 | 1 | 0.30 | 0.25 |
| gpsb151 | 9 | 5 | 3 | 1 | 0.28 | 0.24 |
| Xtxp283 | 4 | 1 | 3 | 0 | 0.33 | 0.27 |
| Xgap342 | 2 | 0 | 1 | 1 | 0.47 | 0.36 |
| Xisep0224 | 11 | 7 | 3 | 1 | 0.26 | 0.23 |
| Xtxp023 | 4 | 1 | 1 | 2 | 0.36 | 0.29 |
| Xiabtp106 | 7 | 4 | 2 | 1 | 0.30 | 0.25 |
| Xiabtp346 | 3 | 0 | 2 | 1 | 0.48 | 0.36 |
| Xiabtp340 | 1 | 0 | 0 | 1 | 0.49 | 0.37 |
| Xiabtp175 | 2 | 0 | 2 | 0 | 0.42 | 0.33 |
| Xiabtp103 | 4 | 0 | 4 | 0 | 0.40 | 0.32 |
| Total | 147 | 59 | 58 | 30 | 11.43 | 9.10 |
| Mean | 5 | 2 | 2 | 1 | 0.37 | 0.29 |
CONCLUSION
The transferability of SSR markers from related families is an alternative approach to bypass the cost developing species specific markers as well as the complexity of the work. The current screened sorghum SSR markers for the purpose of bamboo research will provide a scope of opportunity to researchers for the proper characterization and conservation of bamboo genetic resources. The adequate rate of polymorphism in the markers helps to characterize large number of materials with limited SSR markers. This work will be a foundation particularly in Ethiopia which contributes 67% of the African bamboo area coverage to develop strategies for characterization, and conservation of bamboo genetic resource using molecular approaches. The higher level of rare alleles across both exotic and Ethiopian bamboo species plays an important role in further study of these alleles in detail. Such markers will also guide the implementation of various conservation decisions that will be necessary to ensure that these rare alleles are not lost.
Pair-wise genetic distance between the selected bamboo species.
| Species | G. apus | G. atter | P. ambusoids | G. angostifolia | G. amplexifolia | B. textiles | G. sumatra | A. alpina |
|---|---|---|---|---|---|---|---|---|
| G. atter | 0.35 | |||||||
| P. bambusoids | 0.87 | 0.87 | ||||||
| G. angostifolia | 0.84 | 0.87 | 0.83 | |||||
| G. amplexita | 0.90 | 0.90 | 0.87 | 0.81 | ||||
| B. textiles | 0.81 | 0.85 | 0.84 | 0.82 | 0.84 | |||
| G. sumatra | 0.32 | 0.36 | 0.88 | 0.83 | 0.88 | 0.81 | ||
| A. alpina | 0.80 | 0.87 | 0.86 | 0.80 | 0.81 | 0.83 | 0.80 | |
| O. abyssinica | 0.72 | 0.80 | 0.84 | 0.85 | 0.82 | 0.81 | 0.76 | 0.77 |
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Website along with the published article.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
The expenses of this research were fully covered by the Swedish International Development Agency (SIDA) through Bio-Innovate project “Delivering new sorghum and millets innovations for food security and improving livelihoods in Eastern Africa”- project No. 01/2010. We also gratefully acknowledge the forestry research team of Holetta Agricultural Research Center of the Ethiopian Institute of Agricultural Research for allowing us their bamboo collection garden.


