All published articles of this journal are available on ScienceDirect.
Isolation and Characterization of Amylase Enzyme Produced by Indigenous Bacteria from Sugar Factory Waste
Abstract
Background
Enzymes are biocatalysts commonly used in industries. One of these enzymes is amylase. In Indonesia, most of the enzymes are still imported from overseas. To reduce the production cost, local amylase is needed which can be produced from indigenous microorganisms.
Aims
This research aims to explore isolates amylase-producing bacteria from a liquid waste of sugar factories, optimize amylase enzyme production, and identify genotypically the indigenous bacterial.
Objective
This research aims to explore isolates amylase-producing bacteria from a liquid waste of sugar factories, optimize amylase enzyme production, and identify genotypically the indigenous bacterial.
Methods
This study consists of seven stages: sample preparation, isolation of amylase-producing bacteria, crude amylase extract production, amylase activity assay, optimization of amylase enzyme production, determining the specific activity, and bacterial identification through genotyping.
Results
This research successfully identified 3 bacterial isolates (G-7, G-8, and G-12) that positively produce amylase enzymes from sugar factory waste. The optimal conditions for amylase enzyme production for all three isolates were at 37°C, pH 7.0, and during the exponential growth phase - at 24 hours for isolate G-8 with specific amylase enzyme activity of 0.198 U/mg, and at 48 hours for isolates G-7 and G-12 with specific amylase enzyme activities of 0.108 U/mg and 0.208 U/mg respectively. The 16S rRNA gene identification results showed that G-7, G-8, and G-12 belong to the species Bacillus infantis, Bacillus flexus, and Pseudomonas nitroreducens respectively.
Conclusion
The species Bacillus infantis, Bacillus flexus, and Pseudomonas nitroreducens has shown great potential for the production of amylase enzyme.
1. INTRODUCTION
Enzymes are biocatalysts commonly used in industries. The demand for enzymes in Indonesia reaches 31 thousand tons and continues to increase from 2018 to 2022. However, many of these enzymes aretill imported from China, Turkey, and several other countries [1]. It presents an opportunity to domestically produce indigenous amylase enzymes derived from Indonesian microorganisms to reduce production costs on an industrial scale.
One of the enzymes commonly utilized in industries is the amylase enzyme. Amylase enzyme accelerates the hydrolysis reaction of starch into smaller molecules like dextrin, maltose, and glucose. This enzyme finds widespread application in various industries, including liquid sugar production, ethanol, brewing, textiles, detergents, chemicals, and paper production. In the paper industry, alpha-amylase enzyme is utilized to modify starch by reducing the viscosity of the starch slurry. This modified starch has low viscosity and is subsequently used as a surface coating material in paper, enhancingpaper quality [2].
Amylase enzymes are often found in humans, animals, plants, and microorganisms. However, to meet industrial demand, most amylase enzymes are obtained from microorganisms [3]. The isolation of enzymes derived from microorganisms is chosen due to several advantages when compared to plants or animals. These include rapid microorganism growth, the ability to engineer for recombinant enzyme production, ease of scaling up production, and production conditions unaffected by natural factors such as seasons and time [4]. Several previous studies have succeeded in identifying amylase-producing bacterial species from the genus Bacillus, including Bacillus cereus [5], Bacillus licheniformis [6], Bacillus atrophaeus [7], Bacillus subtilis [8], Bacillus megaterium [9], and Bacillus halotolerans [10] While species from other genus are Pseudomonas balearica [11], Pseudomonas aeruginosa [12], Streptococcus lutetiensis [13] and some others.
On the other hand, Mutlu & Poyraz [14] confirmed that the species Pseudomonas nitroreducens is a bacterium producing amylase enzymes, and Bacillus infantis was confirmed by Saggu & Mishra [15]. However, they have not conducted research on optimizing the incubation time, temperature, and pH of amylase production. Jing W et al. [16] reported amylase production by Bacillus flexus conducted at an incubation time of 10-12 hours at a temperature of 37°C, but no research has been conducted on optimizing the pH of amylase production by Bacillus flexus.
Amylase enzyme-producing bacteria are commonly found in waste and agricultural industrial waste that contains a significant amount of organic waste [17]. In Indonesia, many industries produce agricultural waste, one of which is the sugar factory industry located in the city of Jombang, East Java. The sugar factory industry produces liquid waste with high organic matter content, one of which is starch [18]. The starch contained in the liquid waste of sugar factories can be broken down by bacteria-producing amylase enzymes into simpler molecules.
As of right now, the only publication we could find on the isolation of bacteria from Indonesian sugar industry waste was published in a research conducted by Widyawati [19], which succeeded in identifying three species of bacteria producing amylase enzyme from the liquid waste of Modjopanggoong Tulungagung sugar factory which produced the largest amylolytic index, namely Actinobacillus sp., Pseudomonas stutzeri, and Bacillus cereus. Since the study did not optimize the synthesis of amylase enzyme, bacteria that produce the enzyme from a liquid waste sugar plant located in Jombang, East Java are required to be collected. Variations in the sampling sites may reveal native bacterial species with superior amylase enzyme production capabilities. For even more insight, the synthesis of amylase enzyme must be optimized, and the genotypes of isolates of indigenous bacteria must be identified for further understanding.
2. MATERIALS AND METHODS
The liquid waste samples were collected from a Sugar Factory in Jombang Regency, East Java, Indonesia. All equipment and materials used in this research were funded in 2023 by the Ministry of Education, Culture, Research, and Technology Indonesia.
2.1. Sample Preparation
The liquid waste samples were placed in a sealed bottle and directly put into the ice box. The characterization of samples carried out directly on-site is determined by the temperature and pH of waste samples.
2.2. Isolation of Amylase Enzyme-producing Microbes
The isolation of microbes producing amylase enzymes was carried out using the spread plate technique on a selective media. Starch Agar containing (g/L): Nutrient Agar (NA) 28 and starch 10. Dilution of the sample is carried out by filling 9 test tubes labeled 10-1 to 10-9 with 9 mL of 0.85% NaCl. Then, 1 mL of sample is put into a 10-1 test tube and homogenized. Then, put 1 mL of dilution in a 10-1 test tube into a 10-2 test tube and homogenize. Dilution is carried out to a degree of dilution of 10-9. After that, 100 μL of dilution results were taken from each test tube and then spread on Starch Agar media and incubated at 37°C for 24 hours in the incubator. Then, selected bacterial isolates were to be purified through the strike plate technique.
After obtaining pure isolates, iodine tests were carried out to determine the reaction of starch hydrolysis on Starch Agar media [20]. Bacterial isolates that produce amylase enzymes will produce and secrete extracellular amylase enzymes around the colony so that a clear zone around the colony is visible after rinsing with iodine solution. The clear zone is measured using calipers to calculate the amylolytic index. The formula calculates the amylolytic index according to Eq. (1) [21]:
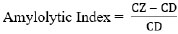 |
(1) |
where CZ was the Clear Zone Diameter (mm), and CD was the Colony Diameter (mm).
2.3. Production of Crude Amylase Extract
One ose of bacterial isolate was put into 20 mL of liquid media containing (g/L): Nutrient Broth (NB) 13 and starch 10, then incubated at 37°C 100 rpm. After 24 hours, 10% of the bacterial culture was taken and put into 30 mL of production media containing (g/L): Nutrient Broth (NB) 13, starch 10, MgSO4.7H2O 0.25, KH2PO4 0.05, CaCl2.2H2O 0.05, MnCl2.4H2O 0.015, and FeSO4.7H2O 0.01. The bacterial growth curve was measured by measuring Optical Density (OD) at 600 nm [22] from 0-168 hours every 24 hours. The culture was centrifuged for 15 minutes at a speed of 10,000 rpm at 4°C to obtain a supernatant, which was then tested for amylase enzyme activity and determination of protein levels.
2.4. Amylase Activity Test
The DNS method measures the concentration of reducing sugars in the supernatant. Glucose solutions with concentration variations of 0, 20, 40, 60, 80, and 100 μg/mL create a standard glucose curve. Determination of reducing sugar concentration in the DNS supernatant method is performed by reacting 1 mL of supernatant with 1 mL of 1% starch solution for 10 minutes. Then, add 2.5 mL of DNS reagent. The mixture reacted at 100°C for 5 minutes. After that, the test tube containing the mixture is cooled to stop the reaction by rinsing it with running water and then ice water so that the test tube does not break. Absorbance was measured at a wavelength of 540 nm [23]. The absorbance value is then fed into a linear regression equation on the standard glucose curve to determine the concentration of reducing sugars in supernatants. The activity of the amylase enzyme is calculated using the formula Eq. (2) [24]:
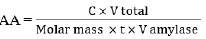 |
(2) |
where AA was Amylase Activity (U/mL), C was the glucose concentration (μg/mL), V was the volume (mL), Molar mass was the Molar mass of glucose (μg/μmol), and t was the incubation time (minute).
2.5. Optimization of Amylase Enzyme Production
The optimization of amylase enzyme production is carried out using the incubation time, pH variations of production media, and incubation temperature using the One Factor at Time (OFAT) method. Incubation time optimization was carried out at a temperature of 37°C [25] and pH 7 [26] for 0-168 hours and taken every 24 hours. The results of optimal incubation time were used for pH and temperature optimization. pH optimization wascarried out at a temperature of 37°C with pH variations of 5, 6, 7, 8, 9, and 10 by adding HCl and NaOH to the desired pH [27]. Incubation temperature variations were carried out at 27, 37, and 47°C using the optimal pH and incubation time obtained previously.
2.6. Determination of Specific Activity
The protein concentration in the supernatant was measured using the Lowry method. Bovine Serum Albumin (BSA) solutions varying in concentration from 0, 50, 100, 150, 200, 250, and 300 μg/mL wereused to create standard protein curves. 0.5 mL of supernatant was reacted with 2.5 mL of Biuret reagent and incubated for 10 min. Then, 0.25 mL of Folin-Ciocalteu reagent was added and incubated for 30 min. Absorbance was measured using a UV-Vis Spectrophotometer at 750 nm wavelength [28]. The absorbance value was then fed into a linear regression equation on the standard protein curve to determine the protein concentration in the supernatant. The formula calculated the specific activity of the amylase enzyme according to Eq. (3) [26]:
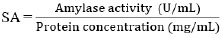 |
(3) |
where SA was the specific activity of the amylase enzyme (U/mg).
2.7. Genotype Identification of Species
The 16S rRNA encoding gene sequencing method was used to identify the resulting bacterial species. DNA isolation was carried out using the ZymoBIOMICS™ DNA Miniprep Kit, then copies of the 16S rRNA encoding gene were multiplied by PCR method using 16s primers (27F-1492R). Then, the quality of the 16S rRNA encoding gene was checked using agarose gel electrophoresis. Furthermore, the 16S rRNA coding gene wassequenced to obtain the nucleotide sequence of the 16S rRNA encoding gene. The results obtained were then analyzed to look for similarities with other bacteria in the gene database contained in NCBI using BLAST [29]. Then, a phylogenetic analysis was carried out to determine the kinship of species with other species.
3. RESULTS
3.1. Isolation of Amylase Enzyme-producing Microbes
Initial characterization of liquid waste from sugar factories obtained a pH of 4.8 and a temperature of 33.8°C. The results of the spread plate with stratified dilution of sugar factory liquid waste samples on Starch Agar selective media that have been incubated for 1x24 hours at a temperature of 37°C, 12 isolates were selected that have the potential to produce amylase enzymes. The 12 isolates came from different dilutions (Table 1). The 12 selected bacterial isolates were then labeled G-1 through G-12.
| Dilution Rate | Number of Bacterial Isolates Taken |
|---|---|
| 10-1 | - |
| 10-2 | - |
| 10-3 | - |
| 10-4 | 7 |
| 10-5 | 2 |
| 10-6 | 2 |
| 10-7 | 1 |
| 10-8 | - |
| 10-9 | - |
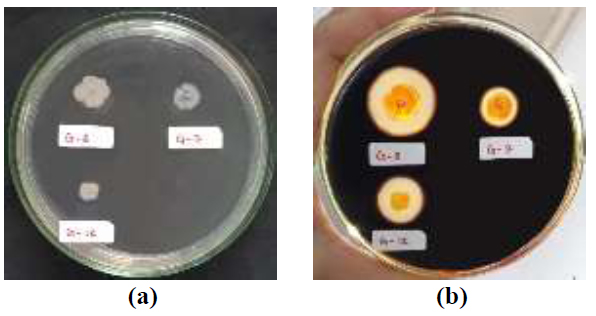
The iodine test results showed that 3 isolates positively produced extracellular amylase enzymes, namely bacterial isolates G-7, G-8, and G-12 (Fig. 1). This is evidenced by the presence of a clear zone around the bacterial colony after rinsing with an iodine solution. The clear zone produced indicates that the starch contained in the Starch Agar media has been hydrolyzed into its monomers by the amylase enzyme produced by bacteria. In contrast, the blue-black dark zone is an iodine-starch complex [20]. Some previous studies have also used iodine tests to screen for amylase-producing bacteria [30-33].
The results showed that the G-12 isolate had the highest amylolytic index, which was 1.29. G-7 and G-8 isolates were 0.38 and 1.14, respectively (Table 2). G-8 isolates have the largest colony diameter when compared to the other two isolates but have a smaller amylolytic index when compared to the G-12 isolate.
| Bacterial Isolate | Colony Diameter (mm) | Amylolytic Index |
|---|---|---|
| G-7 | 10.90 | 0,38 |
| G-8 | 11.08 | 1,14 |
| G-12 | 7.63 | 1,29 |
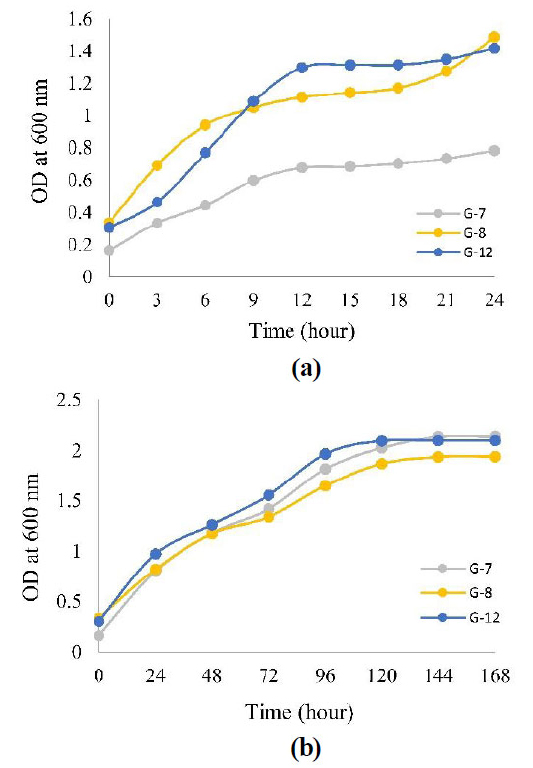
The growth curve of the three isolates was measured every 3 hours for 24 hours, as in Fig. (2a), but the curve still showed an increase in the 24th hour, so measurements were taken every 24 hours for 168 hours, as in Fig. (2b). (Fig. 2a) that even though OD measurements have been measured every 3 hours, the growth curve does not show any phase lag, so it can be concluded that the growth of the three isolates is high speed, which causes the phase lag to be invisible.
3.2. Optimization of Incubation Time, pH, and Temperature of Amylase Enzyme Production
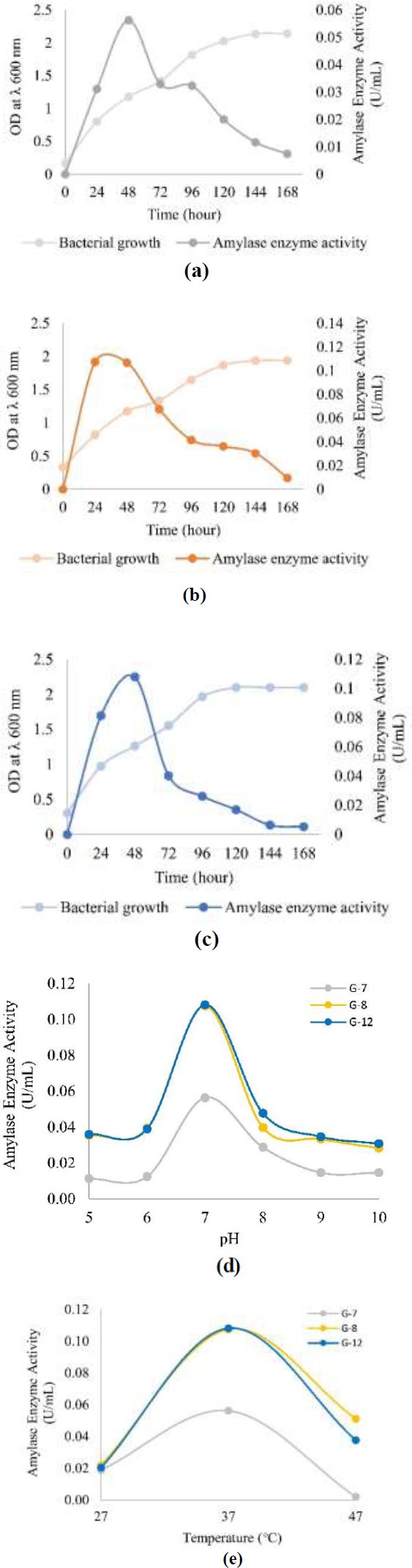
The optimal conditions for producing amylase enzymes in all three isolates were 37°C, pH 7, and in the exponential phase, namely the 24th hour for G-8 isolate with amylase enzyme activity of 0.107 U/mL and the 48th hour for G-7 and G-12 isolates with amylase enzyme activity of 0.056 U/mL and 0.108 U/mL respectively (Fig. 3).
3.3. Determination of the Specific Activity of the Amylase Enzyme
G-12 isolate had the highest amylase enzyme-specific activity, at 0.208 U/mg, followed by G-8 and G-7 isolates at 0.198 U/mg and 0.108 U/mg, respectively (Table 3). The specific activity of an enzyme indicates the number of units of product produced per mg of the enzyme. The higher the value of the specific activity of an enzyme, the greater its purity.
| Bacterial Isolate | Amylase Enzyme Activity (U/mL) | Protein Concentration (mg/mL) | Specific Activity of Amylase Enzyme (U/mg) |
|---|---|---|---|
| G-7 | 0.056 | 0.520 | 0.108 |
| G-8 | 0.107 | 0.542 | 0.198 |
| G-12 | 0.108 | 0.520 | 0.208 |
3.4. Genotype identification of Bacteria
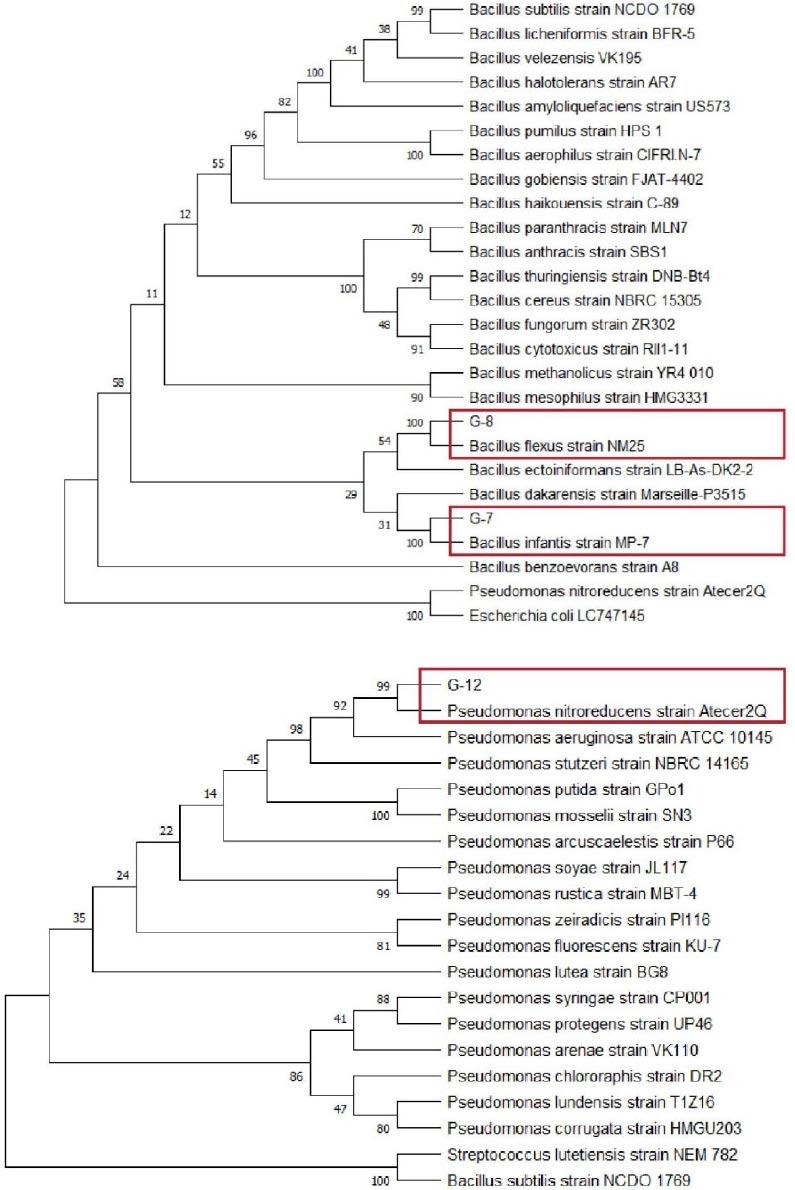
Phylogenetic analysis using MEGA with Neighbor-Joining model showed that G-7 and G-8 isolates belong to the genus Bacillus, while G-12 isolate belongs to the genus Pseudomonas. Fig. (4) shows G-7 isolate closely related to Bacillus infantis species that have a 16S rRNA gene sequence or percentage identity of 99.8%, G-8 related to Bacillus flexus with a percentage identity of 99.9%, and G-12 isolate closely related to Pseudomonas nitroreducens species that have a 16S rRNA gene sequence or percentage identity of 99.7%.
According to Stackebrandt & Goebel [34], if the similarity of the 16S rRNA gene has a “percentage identity” value above 95%, then the similarity is at the genus level, while if the value of “percentage identity” is above 97.5%, then the similarity is at the species level.
4. DISCUSSION
The high demand for enzymes in Indonesia encourages us to look for sources of enzyme producers from within the country. This study seeks to isolate indigenous bacteria from sugar factory waste that can produce extracellular amylase enzyme as a source of amylase enzyme production. The presence of starch in the liquid waste of sugar factories can induce the formation of extracellular amylase enzymes. This starch can be broken down by amylase enzyme-producing bacteria into simpler molecules. This study succeeded in obtaining 3 isolates of bacteria that positively produced amylase enzyme from sugar factory waste. Results that are close to the outcomes of this study have also been brought in by various previous studies [19], but the study has not been optimized for amylase enzyme production.
In the optimum incubation time, results showed that the G-12 isolate had the highest amylase enzyme activity, which was 0.108 U/mL, which occurred at the 48th hour. G-8 isolate had the highest enzyme activity at 24 hours at 0.107 U/mL. Another isolate, G-7, produced the lowest enzyme activity compared to the other two isolates, 0.056 U/mL, at 48 hours. Despite differences in incubation time, all three isolates showed the highest amylase enzyme activity in the exponential phase (Figs. 3a-c). In the exponential phase, bacteria develop rapidly, and there is a drastic increase in the number of bacteria. As a result, the more bacteria there are, the more amylase enzymes are produced. This indicates that the amylase enzyme is used for growth and cell metabolism [35]. Other studies have also shown optimum amylase production in the exponential phase [36], but Deljou [37] reported optimum amylase production in the stationary phase. According to Mubarik et al. [35], this happens because enzymes are used for self-defense or antagonistic response.
After reaching the optimum time, enzyme activity decreases over time (Figs. 3a-c). This is due to the reduced nutrient content in the growth media and the accumulation of toxic compounds [38]. These toxic compounds can be inhibitors that can inactivate the amylase enzyme by covalently binding to a certain group on the active side so firmly that the active side cannot turn the substrate into a product. This is also supported by Nguyen et al. [39] and Raju & Divakar [40], who proved tPseudomonas aeruginosa species also produce α-amylase inhibitors in addition to producing amylase enzymes.
The results showed that at pH close to hábitate pH, which is pH 5, the activity of amylase enzyme isolates G-7, G-8, and G-12, respectively, amounted to 0.011 U/mL, 0.035 U/mL, and 0.036 U/mL. This result is lower when compared to the activity of amylase enzyme at pH 7, which is 0.056 U/mL for G-7 isolate, 0.107 U/mL for G-8 isolate, and 0.108 U/mL for G-12 isolate. Therefore, the optimum pH for producing amylase enzymes from all three isolates is at pH 7, as seen in Fig. (3d). Other studies have also reported that the production of amylase by Bacillus amyloliquefaciens occurs at pH 7 [41].
Although the optimum pH for producing amylase enzymes is pH 7, all three isolates can survive in habitats with acidic environments, pH 4.8. The reason for this could be that bacteria are able to keep cells’ pH levels neutral by eliminating excess H+ ions [42]. In addition, at an alkaline pH, which is pH 8 to 10, the three isolates can also survive but have lower amylase enzyme activity when compared to pH 7 (Fig. 3d). This ability is due to bacteria having Na+/H+ antiporters that can insert H+ ions and emit Na+ ions simultaneously to maintain a neutral pH in the cell [43].
The results showed that at 37°C, all three isolates showed the highest amylase enzyme activity, which was 0.056 U/mL for G-7 isolate, 0.107 U/mL for G-8 isolate, and 0.108 U/mL for G-12 isolate (Fig. 3e). Research conducted by Bukhari & Rehman [25] also reports that bacteria producing amylase enzymes, namely Bacillus subtilis, grow optimally at a temperature of 37°C. The three isolates include mesophile bacteria, which are bacteria that live well at temperatures between 20°C and 45°C [42] so that the three bacteria can live in their natural habitat with temperatures of 33.8°C.
When the surrounding temperature falls below the optimal level, internal chemical reactions and diffusion take longer to complete, slowing down the rate of cell metabolism. In addition, the cell membrane also becomes rigid, which results in the cell membrane losing its fluidity properties [42]. At the same time, at ambient temperatures that are too high from the optimum temperature, the fluidity of the cell membrane increases due to the weakening of interactions between molecules [44]. This can damage cell metabolism due to the entry of molecules that should not enter the cell. In addition, proteins, including enzymes and constituent components of cells, can also be damaged by denaturation [45].
Based on the study, the G-12 isolate had the highest amylase enzyme-specific activity, which was 0.208 U/mg, followed by G-8 and G-7 isolates with amylase enzyme-specific activity of 0.198 U/mg and 0.108 U/mg, respectively (Table 3). The specific activity of an enzyme indicates the number of units of product produced per mg of the enzyme. The specific activity of an enzyme can also determine the purity of an enzyme in a mixture. The higher the value of the specific activity of an enzyme, the greater its purity. The specific activity of the amylase enzyme obtained in this study is lower than the specific activity of the amylase enzyme used in the study of Wang et al. [46], which is >16 U/mg used to modify starch. The starch is then applied as paper surface sizing. The reason for this could be that the amylase enzyme employed in this study is still in the form of crude extract, which makes it possible to purify the enzyme and boost its specific activity.
Phylogenetic analysis based on the 16S rRNA gene sequence showed G-7 isolate to be closely related to Bacillus infantis, G-8 isolate is Bacillus flexus, and G-12 isolate to be closely related to Pseudomonas nitroreducens (Fig. 4). Several studies have shown that some bacterial species of the genus Bacillus and Pseudomonas are bacteria producing amylase enzymes, such as Bacillus cereus and Pseudomonas stutzeri [19], Pseudomonas luteola [47], and Pseudomonas balearica [11]. This study shows that the species Bacillus infantis, Bacillus flexus, and Pseudomonas nitroreducens are bacteria producing the enzyme amylase. Previous research has also confirmed that all three isolates are bacteria-producing amylase enzymes. It is in line with Mutlu & Poyraz [14], who confirmed that the species Pseudomonas nitroreducens is a bacterium producing amylase enzymes, Bacillus infantis was confirmed by Saggu & Mishra [15], and Bacillus flexus was confirmed by Jing et al. [16].
CONCLUSION
Three isolates of amylase-producing bacteria were successfully obtained for this study from the liquid waste of a sugar factory located in Jombang, East Java. The three isolates were identified by 16s rRNA, which revealed that they were Bacillus infantis, Bacillus flexus, and Pseudomonas nitroreducens. The optimal conditions for amylase enzyme production for all three isolates were at 37°C, pH 7, and during the exponential growth phase - at 24 hours for isolate Bacillus flexus with specific amylase enzyme activity of 0.198 U/mg, and at 48 hours for isolates Bacillus infantis and Pseudomonas nitroreducens with specific amylase enzyme activities of 0.108 U/mg and 0.208 U/mg respectively. All three species have shown great potential for the production of amylase enzymes in order to reduce enzyme imports in Indonesia. To meet industrial demand, the specific activity of the amylase enzyme can be increased by purifying the enzyme amylase from crude extract.
LIST OF ABBREVIATIONS
| AI | = Amylolytic Index |
| AA | = Amylase Activity |
| SA | = Specific Activity |
| DNS | = 3,5-dinitrosalicylic Acid |
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article will be available from corresponding author [E.S] upon request.


