All published articles of this journal are available on ScienceDirect.
Signal Peptide Selection for the Efficient Periplasmic and Secretive Expression of Recombinant Brazzein in Escherichia Coli
Abstract
Background:
The high production cost and difficulty of functional expression of brazzein are the limiting factors, making the development of inexpensive, scalable technologies critical for their successful implementation in the market. Secretory expression allows functional expression of the S-S bond-rich proteins and facilitates the purification procedure, resulting in lower processing costs. However, extensive screening and optimization of multiple signal peptides are required to ensure the successful secretion of recombinant proteins.
Objective:
We studied the expression of the minor type of brazzein using 21 different signal peptides in Escherichia coli and investigated their ability to direct the target protein into periplasmic space and culture medium.
Methods:
The synthetic genes were cloned into the pSEVA234 vector under the inducible Trc promoter and initial micro-scale expression analysis was conducted at two distinct conditions followed by scale-up and purification of the selected signal peptides with secretive abilities.
Results:
Two signal peptides led to the secretion of the target protein. The yields of the target protein for MalE_Brazzein and HstI_Brazzein in the periplasm were 11.33 mg/L and 52.33 mg/L, and those in the culture media were 3.975 mg/L and 7.73 mg/L, respectively.
Conclusion:
This study will provide insights into the identification of optimal signal peptides for secretive brazzein expression in E.coli and demonstrate that the abovementioned two signal peptides can be used for successful extracellular production of the target protein in this host.
1. INTRODUCTION
The potential impact of sucrose on glucose homeostasis and its adverse effects are now better understood [1], and the dramatic increase in diabetes and obesity over the past few decades [2] has shifted interest toward ultra-low-calorie, healthful alternatives derived from natural sources. One of these alternatives is sweet proteins, which are up to a few thousand times sweeter than sucrose, do not affect glucose homeostasis, have low energetic value, and are bioengineerable. Brazzein, the smallest known sweet protein to date, was first found in the fruit of Pentadiplandra brazzeana in 1994. Two types of brazzein exist in nature: i) the major form, which has 54 amino acid residues and is 500 times sweeter than sucrose by weight, and ii) the minor form, which lacks the first pyroglutamic acid residue [3]. The latter is two times sweeter than the major form [4]. Brazzein is the most attractive from an industrial standpoint due to its broad pH range, high-temperature stability [3], and sweet taste similar to sucrose [5]. However, the formation of disulfide bonds in recombinant brazzein is crucial for maintaining these properties [4, 6, 7], and yet cytoplasmic expression in E.coli resulted in the formation of inclusion bodies that required further treatment for the recovery of sweet taste [8]. The lack of favorable conditions and mechanisms for the formation of disulfide bonds makes it challenging to obtain the functional form of S-S bond-rich proteins in the cytoplasm of E. coli cells. The periplasm of E. coli, on the other hand, is characterized by an oxidative environment and chaperones involved in the formation and correction of disulfide bonds [9]. Multiple strains have been engineered to form disulfide bonds in the cytoplasm effectively. For instance, researchers reported superior brazzein production in the cytoplasm of T7 SHuffle cells than in BL21 (DE3) strain cells [10]. Another team developed the CyDisCoTM-system by expressing yeast mitochondrial Erv1p and DsbC chaperones using the pLysS backbone in the cytoplasm and implemented it for intracellular brazzein expression [11]. Although this approach allows for the expression of functional recombinant proteins in large quantities, periplasmic or secretive expression offers various advantages, including the ease of purification and the inhibition of protease attack, in addition to accurate folding [12, 13]. In fact, several commercialized Fab fragments and pharmaceutical products at the clinical stage are produced by periplasmic secretion in E. coli [14, 15].
Notwithstanding these benefits and achievements, the limited effectiveness of protein secretion into the extracellular medium in E. coli remains a significant limitation that necessitates the selection of appropriate signal peptides and an extensive optimization process [16]. For instance, Santos, B.D. et al. (2019) successfully enhanced the periplasmic export of small metal-binding protein in E. coli through the screening of different signal peptides [17]. Another team created an antibiotic-based screening approach for optimizing a signal peptide library for periplasmic expression of a single chain variable (scFv) antibody fragment by employing β-lactamase as a C-terminal reporter of periplasmic localization. The method linked scFv translocation to β-lactamase potency and enhanced signal peptides raised periplasmic scFv activity by about 40% [18]. Such technologies greatly expand screening throughput and minimize labour in the selection process. Additionally, robust in silico tools are developed to aid the selection of appropriate signal peptides [19, 20].
In this work, we tried to screen and select different signal peptides suitable for localizing recombinant brazzein in the periplasmic space and directing it to culture media in E.coli. Initially, signal peptides were screened using bioinformatics tools and twenty-one genes conjugated with selected signal peptide coding sequences were designed, and codons were optimized for E.coli to balance codon usage. Synthesized synthetic genes were cloned into the pSEVA234 vector by the restriction-ligation method. The generated constructs were transformed into the E.coli NEB Express strain and microscale expression studies were then carried out under two distinct conditions to evaluate and compare the impact of the selected signal peptides on the synthesis and localization of the target protein. Then, the expression of two selected constructs based on secretive ability was scaled up and recombinant brazzein was purified by nickel affinity chromatography. Finally, the yield of the target protein in the periplasmic space and culture medium was measured.
2. MATERIALS AND METHODS
2.1. BacteRial Strains, Plasmids, Antibodies and Columns
The E.coli strain NEB Express (C2523I) was used as an expression host from NEB Company (USA). The pSEVA 234 expression plasmid used in this work was provided by the SEVA repository (Lorenzo Lab, Spain). For Western blot analysis, 6x-His Tag Recombinant Rabbit Monoclonal Antibody (RM146) from Thermo Scientific (USA) and Anti-Rabbit IgG (A3687)–Alkaline Phosphatase antibody produced in goat (Merck, Germany) were used and stained with NBT/BCIP Stock Solution (Roche Switzerland). Recombinant brazzein was purified using prepacked 1 mL Complete™ His-Tag Purification Columns purchased from Roche (Switzerland).
2.2. Signal Peptide Selection
First, signal peptide sequences suitable for Escherichia coli K-12 strain and some other common bacterial signal peptides were extracted from the signalpeptide.de database. The cutting efficiency of these signal peptides in the production of brazzein protein was evaluated using the SignalP5.0 program that distinguishes diverse signal peptides based on a deep neural network approach [19]. Signal peptides with a high probability of being cleaved at the correct amino acid positions were chosen for the experimental study. While selecting, we also tried to include signal peptides from various pathways. The localization and solubility of the fused recombinant proteins were subsequently analyzed using the ProtComp Version 9 [20] and Protein-Sol [21] software programs, respectively. The ProtCompB server integrates many protein localization prediction methods, including neural network-based prediction and direct comparison with bases of homologous proteins with established localization, and it can predict five possible destinations for Gram-negative bacteria. Protein-Sol scales solubility ranges from 0 to 1 where a value nearest to one being the most soluble. The Protein-Sol server scales solubility from 0 to 1 and interprets the results using the experimental solubility dataset, where any value more than 0.45 is projected to have better solubility than the average soluble E.coli protein [21, 22].
2.3. Designing and Construction Of Expression Vectors
Brazzein sequence was retrieved from the UniProt database (P56552 · DEF_PENBA), and the first amino acid pyroglutamate was removed from the sequence and codon optimized for E.coli using IDTDNA’s codon optimization tool [23]. Further ribosome binding site and codon-optimized signal peptide-coding sequences were added to the N-terminal side, while the 6x His-tag sequence was included in the C-terminus for easy visualization and purification. In addition, to facilitate cloning the genes into pSEVA234 EcoRI and XbaI enzyme sites were added to the 5ʹ and 3ʹ ends of the gene, respectively (Fig. 1). The genes were synthesized, and sequences were confirmed by Synbio Tech (USA).
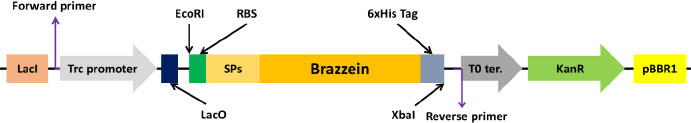
2.4. Transformation and Positive Clone Screening
The prepared constructs were further transformed into NEB Express strain for expression. For this reason, the plasmids were first diluted to a concentration of 10 ng/μl and 20 ng of plasmid DNA was added to 50 µl chemically competent E.coli cells. The transformants were grown on Luria Bertani (LB) agar Petri plates overnight after chemical transformation [24], and for each plasmid, two colonies were picked and separately transferred into 2 mL kanamycin-containing LB medium and grown for approximately 16 h. After this step, part of the culture was subjected to plasmid purification, and glycerol stocks were prepared accordingly.
The presence of the synthetic gene in plasmids isolated from transformed colonies was confirmed by PCR analysis. Q5 polymerase enzyme and buffers (NEB, USA) were used for PCR [25]. Primers designed for the backbone of plasmid pSEVA234 were used: Forward GTTCTGGCAAATATTCTG AAATGAGCTG and Reverse CAAGACTAGTCGCCAGGGT TTTCC. PCR products were analyzed using 1% agarose gel in electrophoresis.
2.5. Protein Expression and Western Blot Analysis
Transformed E.coli NEB Express stocks were thawed at room temperature and a 5 µL stock sample was inoculated into 295 µL kanamycin containing (50 µg/mL) LB medium and grown overnight at 37°C with agitation at 1000 rpm in a 2 mm orbital microplate shaker. Then, 10 µL of this starter culture was transferred to 290 µL LB media (kanamycin: 50 µg/mL) and grown until the OD600 value reached 0.4-0.6 at 37°C with 1000 rpm agitation. Brazzein expression was initiated with 0.5 mM IPTG and conducted under two different conditions: 37°C for 4 hours (h) and 25°C for 16 h. After expression, three types of samples: 1st culture – 50 µL; 2nd supernatant – 100 µL; and 3rd cell pellet from 200 µL culture were collected. All samples were kept at -80°C and used within a week except the culture, which was immediately prepared for electrophoresis by adding equal amounts of 2x Laemmli buffer. The supernatant was first concentrated using a vacuum concentrator before applying for the electrophoresis analysis. The periplasmic expression was checked by extracting proteins through an osmotic shock procedure. The cell pellet was first resuspended in 200 µL of room temperature TES buffer (50 mM Tris-HCl, 10 mM EDTA, and 20% sucrose) and centrifuged at 7 000 x*g for 10 minutes at 4°C. After discarding the supernatant, the pellet was treated with 50 µL of ice-cold TE buffer (50 mM Tris-HCl, 10 mM EDTA) and incubated for 30 minutes on ice, followed by subsequent centrifugation. The taken protein extract was subjected to gel electrophoresis assay accordingly. After running gels, proteins were transferred to nitrocellulose membranes (Cytiva, Sweden), and western blot analysis was performed using the standard methodology [24], diluting antibodies according to the manufacturers’ recommendations. The target protein was visualized by staining with NBT/BCIP substrate for approximately 1 minute.
2.6. Scale-up and Ni AFfinity Chromatography
1 mL of starter culture from two selected constructs was independently transferred to 50 mL of LB media containing 50 g/mL of kanamycin, and protein expression was induced by the addition of 0.5 mM IPTG when the optical density at 600 nm (OD600) reached approximately 0.4-06. Protein synthesis continued for 4 h at 37°C and 300 agitations per minute. The supernatant and cell pellet were then separated by centrifugation and stored at -80 °C until protein purification.
At this stage, to purify brazzein in the periplasm, the previously described osmotic shock process had to be modified to reduce the EDTA-induced chelation of the resin. As a result, an EDTA-free, 50 mM Tris-HCl, pH 8.0 buffer was used as a hypotonic solution. Due to the significant loss in the hypertonic treatment, the two solutions (hypertonic and hypotonic) were combined and subjected to purification after adjusting the NaCl concentration to around 300 mM.
To purify the target protein in the periplasmic extract, first, 10 column volume (CV) buffer A (50 mM Tris-HCl and 300 mM NaCl, pH-8.0) was used to equilibrate the column. Following sample loading, the column was washed with 20 CV of the same buffer. Bound proteins were eluted with 5CV buffer B (250 mM imidazole, 50 mM Tris-HCl, 300 mM NaCl, pH-8.0).
Secreted brazzein was purified without any prior treatment of the culture supernatant following the same purification protocol described above. A schematic view of the experimental design is presented in Fig. (2).
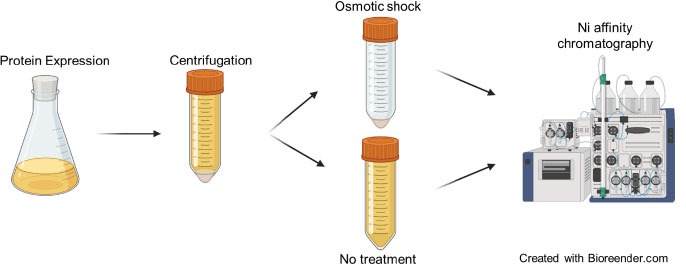
All purification samples were analyzed through 15% Tris-glycine gels [26]. Two parallel gels were run; one was stained with Coomassie R250, while the other was used for western blot analysis. Protein quantification was performed using the Lowry method [27], and the yield of the target protein in the periplasm, supernatant, and cell culture was determined using ImageJ 1.53T [28] based on protein band intensity.
3. RESULTS AND DISCUSSION
3.1. Signal Peptide Selection
As only a few studies have used signal peptides for the periplasmic or secretive brazzein expression in E.coli [29, 30] we decided to use 21 signal peptides to study their ability to direct recombinant brazzein into the periplasm and extracellular space (Table 1). Most of the selected signal peptides belonged to the general secretion (Sec) pathway (17 Sec-dependent-posttranslational pathways and 2 signal recognition particle-dependent -cotranslational pathways), whereas only two were related to the twin-arginine translocation (TAT) pathway. In fact, Sec is predicted to transport more than 90% of all secreted proteins in E.coli [31]. For this reason, signal peptides belonging to the Sec system, along with TAT pathway representatives are widely used in recombinant protein expression [12, 16]. Both pathways, however, belong to the type II secretion system, which transports proteins in two phases: periplasmic translocation and extracellular transport. In the Sec pathway, the cytosolic chaperone SecB or signal recognition particle recognizes an unfolded preprotein and transports it to the corresponding inner membrane-bound protein. The TAT system, in contrast, recognizes a folded preprotein by its complex. In both cases, signal sequences are removed before the protein being released into the periplasm [32, 33]. Mechanisms such as T2SS and T5SS may then transport the proteins from the periplasm to the extracellular environment [34].
Signal peptidases with active sites in the periplasm cut signal peptides before translocation of extracellular proteins; however, if signal peptidases fail to cut signal peptides, the protein will be inactive even if it can pass through the membrane or will end up in the wrong location in the cell [35]. The cleavage efficiency of the selected signal peptides from the correct amino acid residue (Signal peptide - DK...) ranged from 92.24 to 99.78%, indicating a high likelihood of proper processing of the target protein. The signal peptides BglX, FhuD, OmpW, and TorZ were more likely to perform this cut, whereas MalE was the least probable.
In addition, for the selected experimental set, the predicted solubility using the protein-sol platform (Table 1), all sequences were soluble values ranging from 0.513 to 0.758. The highest solubility was observed for PhoA and OmpF meanwhile, FhuD, TorZ and PelB showed the lowest solubility values, though all were still greater than the mean solubility value (0.45), as indicated by Niwa et al. [22]. Because insoluble proteins can accumulate as inclusion bodies, secretory protein solubility may be regarded as the most significant factor in secretion [36]. This criterion did not appear to be a constraining characteristic in our investigation; based on the solubility results of all selected constructs, all of them were chosen for mini-scale expression. The localization prediction, on the other hand, suggested diverse destinations: cytoplasm for three of the constructs, periplasmic space for eight, outer membrane for three, and culture medium for seven.
3.2. Generation of Vector Constructs, Transformation and Selection of Positive Clones
Plasmid pSEVA234 is a vector with 4550 nucleotide pairs and a set of conventional restriction sites enabling easy substitution of its constituent elements [37]. Since the plasmid lacked the RBS site for efficient expression of the target protein under the trc promoter, the RBS motif commonly used for E.coli AGGAGG was inserted into the 5' end of the gene [38]. Previously, 5-8 nucleotides between the RBS and the start codon resulted in considerably more efficient protein expression [39]. As a result, 6 nucleotides were included between the RBS and the start codon of the genes used in this study.
| - | Construct Name | SP Amino Acid Sequence | Pathway Type | Cleavage Site and Probability | Predicted Localization | Predicted Solubility |
|---|---|---|---|---|---|---|
| 1. | PelB_Brazzein | MKYLLPTAAAGLLLLAAQPAMA | SEC | AMA-DK. Pr: 0.9942 | Cytoplasmic | 0.572 |
| 2. | PhoA_Brazzein | MKQSTIALALLPLLFTPVTKA | TKA-DK. Pr: 0.9626 | Cytoplasmic | 0.641 | |
| 3. | OmpA_Brazzein | MKKTAIAIAVALAGFATVAQA | AQA-DK. Pr: 0.9725 | Periplasm | 0.696 | |
| 4. | OmpC_Brazzein | MKVKVLSLLVPALLVAGAANA | ANA-DK. Pr: 0.9561 | Secreted | 0.663 | |
| 5. | OmpW_Brazzein | MKKLTVAALAVTTLLSGSAFA | AFA-DK. Pr: 0.9966 | Secreted | 0.691 | |
| 6. | OmpF_Brazzein | MMKRNILAVIVPALLVAGTANA | ANA-DK. Pr: 0.9899 | Secreted | 0.718 | |
| 7. | LivK_Brazzein | MKRNAKTIIAGMIALAISHTAMA | AMA-DK. Pr: 0.9892 | Periplasm | 0.682 | |
| 8. | PhoE_Brazzein | MKKSTLALVVMGIVASASVQA | VQA-DK. Pr: 0.9897 | Secreted | 0.758 | |
| 9. | MalE_Brazzein | MKIKTGARILALSALTTMMFSASALA | ALA-DK. Pr: 0.9224 | Periplasm | 0.684 | |
| 10. | Btub_Brazzein | MIKKASLLTACSVTAFSAWA | AWA-DK. Pr: 0.9901 | Secreted | 0.708 | |
| 11. | FecA_Brazzein | MTPLRVFRKTTPLVNTIRLSLLPLAGLSFSAFA | AFA-DK. Pr: 0.9864 | Outer membrane | 0.603 | |
| 12. | HstI_Brazzein | MKKNIAFLLASMFVFSIATNAYA | AYA-DK. Pr: 0.9735 | Secreted | 0.684 | |
| 13. | UidC_Brazzein | MRKIVAMAVICLTAASGLTSAYA | AYA-DK. Pr: 0.9574 | Secreted | 0.665 | |
| 14. | BglX_Brazzein | MKWLCSVGIAVSLALQPALA | ALA-DK. Pr: 0.9978 | Cytoplasmic | 0.627 | |
| 15. | PotD_Brazzein | MKKWSRHLLAAGALALGMSAAHA | AHA-DK. Pr: 0.9945 | Periplasm | 0.681 | |
| 16. | OstA_Brazzein | MKKRIPTLLATMIATALYSQQGLA | GLA-DK. Pr: 0.9330 | Outer membrane | 0.706 | |
| 17. | IutA_Brazzein | MMISKKYTLWALNPLLLTMMAPAVA | AVA-DK. Pr: 0.9760 | Outer membrane | 0.583 | |
| 18. | TolB_Brazzein | MKQALRVAFGFLILWASVLHA | SRP | LHA-DK. Pr: 0.9752 | Periplasm | 0.633 |
| 19. | DsbA_Brazzein | MKKIWLALAGLVLAFSASA | ASA-DK. Pr: 0.9672 | Periplasm | 0.698 | |
| 20. | TorZ_Brazzein | MTLTRREFIKHSGIAAGALVVTSAAPLPAWA | TAT | AWA-DK. Pr: 0.9963 | Periplasm | 0.519 |
| 21. | FhuD_Brazzein | MSGLPLISRRRLLTAMALSPLLWQMNTAHA | AHA-DK. Pr: 0.9970 | Periplasm | 0.513 |
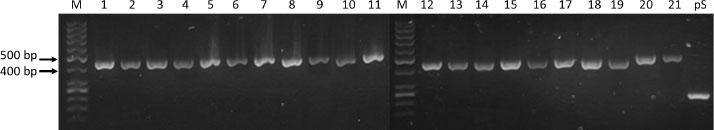
Transformation of the generated constructs into E.coli NEBExpress strains gave >20 colonies when selected on agar plates based on their ability to grow in an antibiotic medium. PCR analysis from purified plasmids that were extracted from picked colonies confirmed the presence of PCR products between 400 and 500 base pair DNA markers in brazzein constructs, which corresponds to the length of the inserted DNA fragments (Fig. 3).
3.3. Identification of Efficient Signal Peptides for Recombinant Brazzein Expression
High-throughput screening techniques are especially desirable because they expedite the research process and reduce screening expenses. Thus, at this stage, a mini-expression study was conducted for the initial evaluation of the secretive ability of the constructs. In addition, since temperature affects the localization of recombinant proteins in the periplasm and culture media, the expression is conducted under two distinct high and low-temperature conditions. Due to the comparatively rapid expression rate of E.coli cells at 37°C, the recommended expression period is between 2-4 h. In contrast, expression at lower temperatures may be slower, necessitating additional time for optimal target protein output [40, 41]. As a consequence, the level of protein expression was evaluated under two distinct conditions. In the first set of experiments, expression was carried out at 37°C for 4 h, while in the second set, expression was carried out at 25°C for 16 h. The evaluation of protein expression from three sample points: i) culture, ii) supernatant, and iii) periplasm are presented in (Table 2).
Table 2.
| - | Construct Name | 37°C 4 h Induction | 25°C 16 h Induction | ||||
|---|---|---|---|---|---|---|---|
| Culture | Supernatant | Periplasm | Culture | Supernatant | Periplasm | ||
| 1. | PelB_Brazzein | ++ | - | - | + | - | - |
| 2. | PhoA_Brazzein | ++ | - | + | + | - | - |
| 3. | OmpA_Brazzein | + | - | - | + | - | - |
| 4. | OmpC_Brazzein | - | - | - | + | - | - |
| 5. | OmpW_Brazzein | - | - | - | + | - | - |
| 6. | OmpF_Brazzein | - | - | - | + | - | - |
| 7. | LivK_Brazzein | - | - | - | + | - | - |
| 8. | PhoE_Brazzein | - | - | - | + | - | - |
| 9. | MalE_Brazzein | +++ | - | + | ++ | + | + |
| 10. | Btub_Brazzein | +++ | - | + | ++ | - | + |
| 11. | FecA_Brazzein | - | - | - | - | - | - |
| 12. | HstI_Brazzein | ++++ | + | + | +++ | + | ++ |
| 13. | UidC_Brazzein | + | - | - | + | - | - |
| 14. | BglX_Brazzein | + | - | - | + | - | - |
| 15. | PotD_Brazzein | - | - | - | - | - | - |
| 16. | OstA_Brazzein | + | - | - | + | - | - |
| 17. | IutA_Brazzein | - | - | - | - | - | - |
| 18. | TolB_Brazzein | - | - | - | - | - | - |
| 19. | DsbA_Brazzein | ++ | - | - | - | - | + |
| 20. | TorZ_Brazzein | ++ | - | - | ++ | - | + |
| 21. | FhuD_Brazzein | + | - | - | - | - | - |
| 22. | pSEVA | - | - | - | - | - | - |
12 constructs expressed brazzein at 37°C compared to 16 constructs at 25°C. In the first set, four signal peptides resulted in the localization of the target protein in the periplasm, while only HstI led the protein to the nutrient medium. However, when the expression was conducted at a lower temperature, the target protein was localized in the periplasm in 5 constructs and in the supernatant in HstI and MalE constructs. For four signal peptides, the FecA, PotD, IutA and TolB, no detectable level of expression was found in both sets. Hence, a greater proportion of constructs demonstrated detectable expression at the lower temperature, the yield of the target protein was significantly greater at 37°C than at 25°C. The signal peptides PhoA, MalE, BtuB, HstI, FhuD and TorZ could successfully direct recombinant brazzein into periplasmic space, though the localization prediction was cytoplasmic for PhoA_Brazzein. The localization prediction for the secretive signal peptides MalE and HstI were periplasmic and secretive, respectively. Interestingly, MalE had the least cutting probability but still could transport the protein into the medium. As a result, MalE_Brazzein and HstI_Brazzein, the two constructs with secretory ability, were chosen for larger-scale expression and purification.
It was also found that the molecular weight of the target protein in both periplasm and culture supernatant was less than that of the 10-kDa protein standard. This was consistent with the estimated molecular mass of the target protein (7.2 - 7.4 kDa following signal peptide cleavage). In some constructs, we also observed the presence of two bands corresponding to the target protein and its precursor, indicating partial maturation of the target protein. In addition, we witnessed that because brazzein contains four disulfides, the protein had a tendency to aggregate in SDS gel electrophoresis regardless of treatment with beta-mercaptoethanol.
3.4. Larger-scale Expression of the Selected Constructs and Purification by Ni Chromatography
The ability of the histidine amino acid to exhibit a strong affinity for metal ions, such as nickel (Ni), copper (Cu), and cobalt (Co), is extensively employed in the process of purifying recombinant proteins. The tag can be conjugated at either terminus of the protein, and it generally does not impede the normal functioning of the protein [42]. Purification of the target protein through a single chromatographic stage yielded acceptable purity for both periplasmic and supernatant samples (Fig. 4).
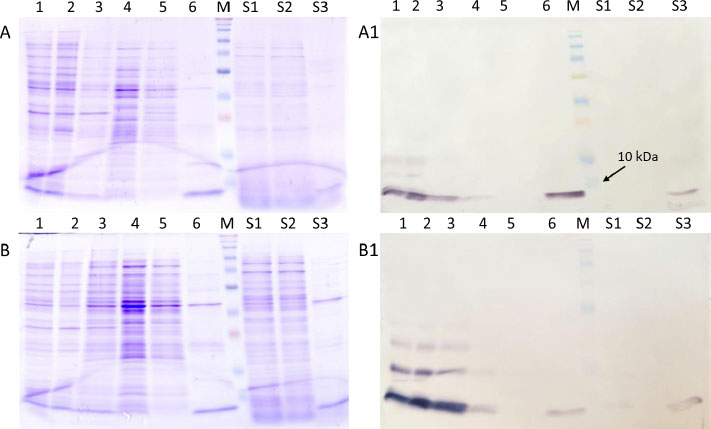
The results suggested that the majority of the target protein remained in the cell pellet following the osmotic shock process (Fig. 4), Line 2). We did not, however, determine if this protein was localized intracellularly (rather than being transported to the periplasm) or if it was in the periplasm and was not released during the osmotic shock process. Therefore, we assume that optimizing the osmotic shock process may increase protein yield, as Ghamghami E et al. (2020) demonstrated that optimizing the four independent variables that influence the TES osmotic shock method could increase the recovery of the anti-G17-Gly scFv antibody fragment [43].
In the samples isolated from the culture supernatant, we observed that the target protein was almost not observed in flowthrough (Fig. 4), Line S2) and wash fractions. This shows that although the nutrient medium was not adjusted before purification, the binding property of the target protein to Ni metal was not affected.
The results showed that the target protein was more abundant in the periplasm than in the culture supernatant. The amount of secreted target protein for MalE_Brazzein and HstI_Brazzein was 3.975±1.5 7.73±2.1 mg/L, respectively (Table 3).
Brazzein is produced intracellularly in E.coli in multiple studies [8, 10, 44, 45]. According to these reports, yield ranged from only several milligrams to approximately 150 mg/L, and the sweetness of brazzein differentiated from not sweet to a similar level of sweetness as wild-type brazzein. We, however, decided to omit sweetness analyses in our study because a prior study reported that recombinant brazzein with C-terminal his-tag lacked sweetness [10]. Additionally, it has been demonstrated that the presence of an intact C-terminus plays a crucial role in the determination of sweet taste [4].
| Names | Extracted Periplasmic Brazzein (mg/L) | Secreted Brazzein Yield (mg/L) |
|---|---|---|
| MalE_Brazzein | 11.33±2.9 | 3.975±1.5 |
| HstI_Brazzein | 52.33±7.6 | 7.73±2.1 |
On the other hand, only a few studies have investigated the direction of brazzein into the periplasm or the extracellular space in E.coli. For example, Lee et al. (2010) employed the pelB signal peptide and a sophisticated gene optimization technology for brazzein expression and showed that the yield of target protein in the periplasmic space could reach 6.7 mg/L but no other signal peptides were covered [29]. There is also a patent in which the brazzein yield claimed to be from 0.5 g to 5.0 g, where authors used 22 different signal peptides for brazzein expression in the BL21 (DE3) strain [30]. However, their ability to localize recombinant brazzein into different compartments of E.coli was not clarified.
Similar research assessed and characterized the efficacy of brazzein secretion with seven distinct signal peptides in Pichia pastoris and brazzein expression levels varied between 283 mg/L to 345 mg/L for the three most successful signal peptides [46]. The reason for such high yield expression could be the advanced secretion system exhibited by yeasts. On the contrary, the expression of proteins in the secretory pathway of E.coli bacteria is typically characterized by low production yields. Successful cases have only been achieved through the implementation of carefully optimized expression systems, high-density fermentation techniques, appropriate environmental conditions, and other strategic approaches [47-49]. For this reason, we expect optimizing other parameters using similar strategies will significantly increase brazzein secretion.
CONCLUSION
Signal peptides suitable for secretory or periplasmic expression of the brazzein gene in E.coli were screened. Among the selected 21 signal peptides, brazzein conjugated with MalE, BtuB, and HstI signal peptides exhibited the highest levels of brazzein expression. However, only two signal peptides, MalE and HstI, have demonstrated the ability to effectively guide brazzein into the culture media with a target protein yield of 3.975 mg/L and 7.73 mg/L, respectively. The findings can help optimize the expression of recombinant brazzein in E.coli cells and contribute to the development of strategies for targeting proteins. However, in order to develop a feasible technology for large-scale production, it is necessary to include additional optimization strategies.
AUTHORS' CONTRIBUTIONS
MM and TS designed the study, MM, EK and SK conducted the experiments. MM, DD and TS wrote and edited the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The study has been funded by the Science Development Fund of the Ministry of Higher Education, Science and Innovation of the Republic of Uzbekistan within the framework of financing practical projects of young scientists. Project number A IRV – 2021-468.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
This project was financed by the Science Development Fund of the Ministry of Higher Education, Science and Innovation of the Republic of Uzbekistan within the framework of financing practical projects of young scientists.
We also thank the founders and contributors of the SEVA collection for sharing the pSEVA234 plasmid with us at no cost.


