All published articles of this journal are available on ScienceDirect.
A Mini-review on Oncolytic Newcastle Disease Virus (NDV): From Highly Contagious Virus to a Biological Tool for Cancer Therapy
Abstract
Newcastle disease virus is a highly contagious viral infection affecting a plethora of avian species with distinct levels of susceptibility. It exerts a significant economic impact in certain countries due to its pathogenic nature, causing high mortality and morbidity rates. It is well characterized that the Newcastle disease virus is among the avian paramyxovirus serotypes, which could be easily disseminated through contaminated feed, water, and others. In view of its capability to thrive in extreme conditions, the exploration of Newcastle disease virus, as an oncolytic agent, has been gaining interest over the last few years. It is widely utilized as a vector in vaccine development for both humans and animals. The versatility in transcription, low deoxyribonucleic acid phase during replication, as well as low recombinant frequency makes Newcastle disease virus a major reason in the development of cancer vaccines. This review highlights the current understanding of its biology, associated with advanced molecular biology tools as oncolytic agents. Given that Newcastle disease virus is still in the early stage of clinical trials as oncolytic agents, deeper exploration of preclinical studies is necessary to ensure its safety and efficacy.
1. INTRODUCTION
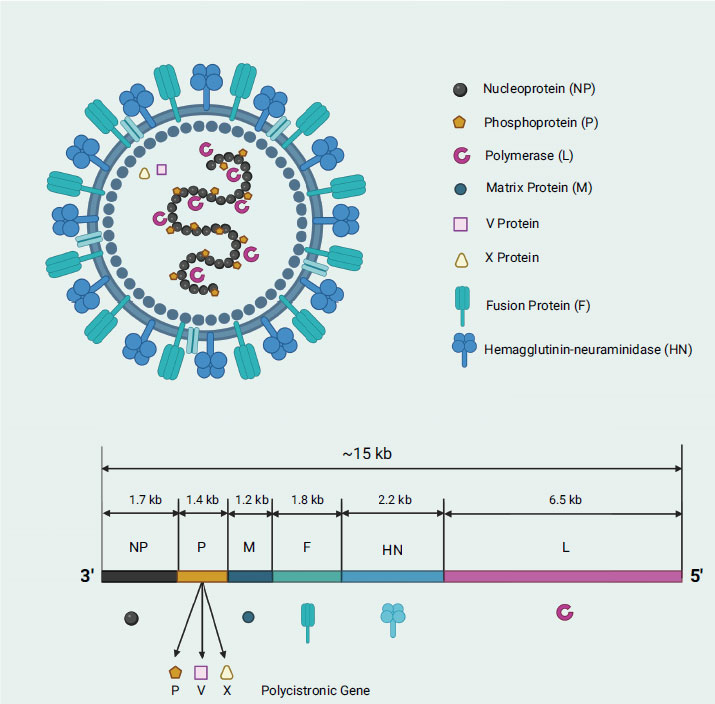
Newcastle disease virus (NDV) is a well-known and prevalent poultry disease in the world as it infects a variety of birds, including chicken, ducks, geese, and turkey, causing economic loss worldwide (i.e., ranked the third most-significant poultry disease with 109 member countries of World Organization for Animal Health (OIE) affected) [1, 2]. NDV was named after the city of Newcastle-upon-Tyne, England, where the first outbreak was reported in the early 1900s. NDV, classified under the Paramyxoviridae family (i.e., like measles virus, mumps virus, respiratory syncytial virus, and parainfluenza viruses), is a type 1 avian paramyxovirus (APMV-1), which is spread through contaminated feed, water, or equipment as well as through direct contact with infected birds or their secretions [3-7]. In severe circumstances, infected birds may die unexpectedly with no apparent symptoms of illness. NDV is an enveloped virus that falls into two classes based on its genome: class I viruses with a genome size of 15,198 nucleotides, and class II viruses with a genome size of either 15,186 or 15,192 nucleotides [8]. NDV, like other viruses, abides by the “rule of six,” which dictates that the number of nucleotides present is always a multiple of six [9]. The non-segmented, single-stranded, negative-sense ribonucleic acid (RNA) genome contains six open reading frames (ORF) which encodes six essential genes in the order of 3’-NP-P-M-F-HN-L-5’ expressing main structural proteins namely, nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin-neuraminidase (HN), and RNA-dependent RNA polymerase (L) (Fig. 1) [10]. The nucleocapsid protein plays a role in binding with the viral RNA, forming the ribonucleoprotein (RNP) complex, while the phosphoprotein is critically important for viral RNA synthesis and acts as a cofactor (i.e., structural bridge that connects the L polymerase and nucleocapsid proteins) for the L polymerase transcription and replication activities. The phosphoprotein consists of three domains: 1) N-terminal domain (NTD), whereby its complexes formation with nascent N protein monomer (N0) prevents its random encapsulation of non-specific RNA; 2) central oligomerization domain (OD), which stabilizes the L protein in the L-P complex; and 3) C-terminal X domain (XD), which interacts with the RNPs to facilitate the entry of template RNA into the template entry channel, and also aids the entry of NTPs into the NTP entry channel [11]. Other than that, matrix protein is vital for virus assembly and budding, while fusion protein mediates viral entry into host cells. On the other hand, the hemagglutinin-neuraminidase protein is involved in virus attachment, entry, and release from host cells, whereas polymerase is responsible for viral RNA synthesis and replication [12]. Interestingly, NDV L polymerase, which possesses four vital channels (template entrance, template exit, product exit, and NTP entrance), is the last gene transcribed during the viral replication cycle [11]. Of note, NDV also has two non-structural proteins, namely, V and W proteins, which are generated by RNA editing during P gene transcription [13]. It is intriguing that the conserved editing site (UUUUUCCC) in the genome sense undergoes the insertion of one or two G residues to generate the V mRNA (which has a +1 frameshift) and W mRNA (which has a +2 frameshift), resulting in a proportion of 68%, 29%, and 2% for the P/V/W protein-encoding mRNAs, respectively [8, 14]. It is worth mentioning that the P, V, and W proteins have identical amino terminals (N-terminal region) but differ in length and amino acid composition at their carboxyl terminus (C-terminal region) [15].
2. PATHOTYPIC CHARACTERIZATION OF NDV
NDV is known for its genetic diversity, with a high mutation rate and frequent recombination events leading to the emergence of new NDV strains with altered virulence or host range [16]. NDV strains can be categorized into three different pathotypes, namely lentogenic (low virulent), mesogenic (intermediate), and velogenic (high virulent), whereby their virulence classification is based on the amino acid sequence of the fusion (F) protein cleavage site, with velogenic strains typically having a multi-basic amino acid sequence at positions 112-116 and a phenylalanine at position 117 nucleotides. In comparison to the velogenic NDV strains, lentogenic strains have a mono-basic cleavage site at positions 112-116 and leucine residue at position 117 nucleotides [8, 17-19]. The intracellular furin-like proteases present in avians’ tissues would cleave the amino acids at those positions (i.e., required for virus maturation and infectivity). Apart from the chemistry of F0 cleavage site for NDV rapid pathotyping (i.e., molecular-based assays), viral isolates are frequently categorized into velogenic, mesogenic, and lentogenic strains based on pathogenicity indices, such as the mean death time (MDT) observed in 9-10-day-old embryonated chicken eggs, and the intracerebral pathogenicity index (ICPI) measured in 1-day-old chicks [20]. Nevertheless, it is important to note that the pathogenicity of NDV strains can vary depending on the host species, and different NDV strains can cause different levels of disease in different bird species [21]. Therefore, the pathogenicity of NDV strains cannot be solely determined by their classification into subtypes. The lentogenic strains of NDV are typically linked with subclinical disease characterized by mild respiratory symptoms such as coughing, gasping, sneezing, and rales (i.e., asymptomatic enteric disease). However, if there are other co-infectious agents present, it can lead to more severe symptoms [22]. On the other hand, mesogenic strains of NDV are intermediate in severity, which can cause acute respiratory disease, and neurological symptoms in certain species. Despite the mortality rate is generally low, usually less than 10%, the infected birds may exhibit signs of nervous system dysfunction such as incoordination and paralysis [8, 23]. Among these, the velogenic strain is regarded as the most fatal as it causes severe respiratory and nervous symptoms and high mortality [24]. Early clinical indications may vary, but can include lethargy, loss of appetite, ruffled feathers, swelling, and redness of the conjunctiva. As the illness advances, affected birds may experience greenish or white watery diarrhea, difficulty breathing, inflammation of the head and neck, often accompanied by bluish discoloration, haemorrhagic gastroenteritis, pneumonia, and encephalitis [20, 24]. Of note, some strains can also cause conjunctivitis in humans who encounter with the infected birds [10].
3. GENOTYPING OF NDV
Thanks to molecular techniques such as PCR, sequencing, and phylogenetic analysis, the NDV strains can be grouped into different genetic lineages, with some lineages being associated with specific geographic regions or host species [2]. The genetic diversity of NDV strains is reflected in their classification into different subtypes based on differences in the hemagglutinin-neuraminidase (HN) and fusion (F) proteins [25]. It is worth mentioning that NDV is divided into two classes of genotypes: class I strains, which have mainly been isolated from wild and domesticated birds found in Africa, Asia, Europe, and America (mostly are low-virulence strains) and class II NDV, comprises a blend of viruses with varying levels of virulence that lead to outbreaks in various regions across the globe [26, 27]. Class I NDV strains are categorized into a singular genotype and three sub-genotypes, whereas class II NDV strains are classified into no fewer than 20 different genotypes (I-XXI) consisting of multiple sub-genotypes (Table 1). According to the classification rules established by Diel and his colleague, if the evolutionary distance between clades within a genotype falls within the range of 3% to 10%, then it is possible to divide that genotype into sub-genotypes [28]. For example, within genotype VII, there are three sub-genotypes, whereas several sub-genotypes exist within genotypes I, V, VI, VII, XII, XIII, XIV, and XVIII [29]. Of note, the Queensland V4 and Ulster/chicken/ Ireland/1967 vaccine strains belong to genotype I whereas LaSota and B1 strains, which are predominantly found in domestic fowl, chicken, and wild birds across North and South America, Africa, Asia, and Europe belong to genotype II [30, 31]. Meanwhile, the mesogenic Mukteshwar strain, which is a well-known vaccine strain, belongs to genotype III isolates where these isolates were first obtained from birds in Japan in the 1930s and later in Pakistan in the mid-1970s before reappeared once more in China less than twenty years ago [32, 33]. Other than that, the genotype IV isolates, which comprise the Herts/33 strain, were found in European poultry before the 1940s [34]. On the other hand, genotype V isolates first appeared in America around the 1970s and later spread to Europe in the 1980s and currently there are cases reported in East Africa, indicating that their geographic distribution is expanding, also suggesting that these isolates are continuously evolving [35, 36].
| Genotypes | Sub-genotypes | Example of NDV Strains | References |
|---|---|---|---|
| I | Ia, Ib, Ic | BHG/Sweden/94, Ulster, V4 | [20, 37] |
| II | - | LaSota, Hitchner B1 | [38, 39] |
| III | - | Mukteshwar | [40] |
| IV | - | Herts/33 (UK) | [41] |
| V | Va, Vb, Vc, Vd | Anhinga (US) | [20] |
| VI | VIa, VIb, VIc, VId, VIe, VIf, VIg, VIh, VIi, VIj, Vik | Pigeon paramyxoviruses | [42] |
| VII | VIIa, VIIb, VIIc, VIId, VIIe, VIIf, VIIg, VIIh, VIIi | Banjarmasin/010, IBS002 | [43, 44] |
| VIII | - | AF22440-i | [45] |
| IX | - | F48E8, duck/China/Guangxi19/2011 | [46] |
| X | - | Chicken_USA_TX_GB_1948 | [29] |
| XI | - | HQ266602 chicken MG 72508, JX18875/2009/MGBBS | [47] |
| XII | XIIa, XIIb XIId | poultry/Peru/1918-03/2008, Goose/Guangdong/2010, NCXMT/Vietnam/2014, NDV15A1 | [48, 49] |
| XIII | XIIIa, XIIIb, XIIIc | KY828160MZ-6/chicken/India/2016, GU585905 chhicken/Sweden/1977 | [50] |
| XIV | XIVa, XIVb | Niger/06 | [51] |
| XV | - | Originated from mixed virulent and vaccine viruses | [20] |
| XVI | - | Mexico_Queretaro_452_1947 | [52] |
| XVII | XVIIa, XVIIb | BF/08 | [51] |
| XVIII | XVIIIa, XVIIIb | ML57051T/chicken/Mali/2010, ML038_07/guinea fowl/Mali/2007 | [50] |
| XIX | - | Cormorant_USA_FL_41105_2012 | [52] |
| XX | - | AF458018_chicken_China_Sh_2_1998, KU373026_ostrich_China_SX_01_2006 | [53] |
| XXI | - | Pigeon-derived NDV isolate pigeon/Bangladesh/BD-P01/2010 | [53] |
4. NDV INFECTION AND REPLICATION INSIDE HOST CELL
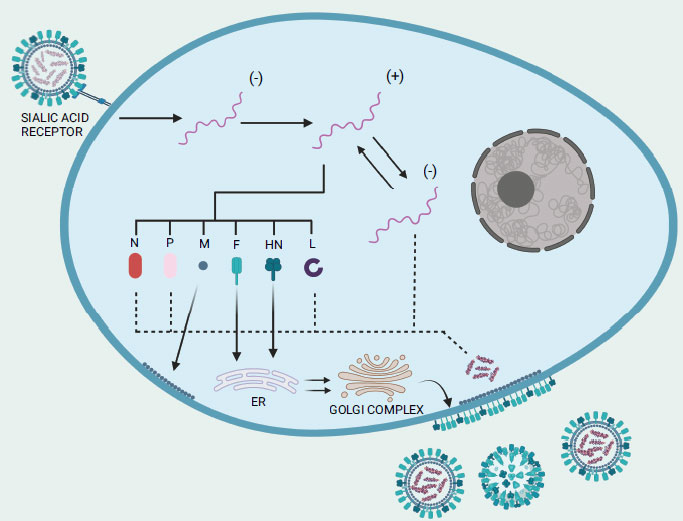
Before infecting and replicating inside the host target cells, the virus binds to the cell surface through two viral surface proteins: a 74 kDa haemagglutinin-neuraminidase (HN) protein and a 67 kDa fusion (F) protein (Fig. 2) [54]. The lectin-like cell binding domain of the HN protein attaches to the α2-3 and α2-6-N-linked sialic acids (i.e., carbohydrate side chains) of the host cell surface receptors and subsequently, proteolytic cleavage activity triggers the conversion of the inactive precursor F0 of the fusion protein into the active disulfide-linked chains F1 and F2 [55]. Shortly after the viral membrane fuses with the host cell membrane, the viral RNA-genome in the negative strand form is introduced into the cytoplasm of the host cell before it undergoes transcription to produce messenger RNAs, which are then translated into viral proteins [56]. Of note, during the transcription process, the L protein also performs modification on the newly synthesized viral mRNA such as 5′ capping, methylation, and polyadenylation [57]. It is important to note also that once a sufficient amount of viral proteins has accumulated, viral transcription transitions to viral replication [58]. The polymerase complex produces a complete plus-strand antigenomic RNA that acts as a template for the synthesis of minus-strand genomic RNA [58]. After undergoing post-translational modifications, the envelope proteins HN and F, along with the M protein, migrate to the host cell membrane. There, they facilitate the encapsulation of a single copy of the NDV genome. Now, the complete progeny virus structure can be released from the infected cell by budding process [54]. Neuraminidase activity of the HN protein mediates the detachment of the virus from the cell as well as facilitates in the elimination of sialic acid residues from the progeny virus particles to prevent self-aggregation [8].
5. NDV EVASION FROM IMMUNE SYSTEM
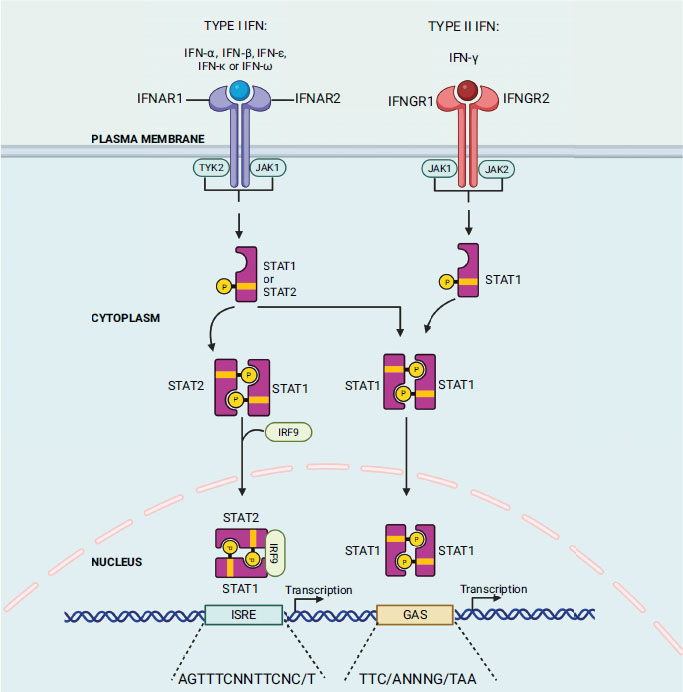
The host body produces interferon (IFN) as the primary cytokine in response to external pathogens such as viruses, bacteria, and mitogens. Previous studies also indicate its involvement in controlling tumour cells and autoimmunity [59]. Isaacs and Lindenmann first discovered interferon in 1957 by observing that supernatants from virus-infected cells contained a protein/factor that confers resistance to the virus on other uninfected cells [60]. Interferon was named for its capacity to impede virus growth. At present, interferon is categorized into three subtypes: type I family, consisting of IFN-α, β, ε, κ, and ω; type II family, IFN-γ; and type III IFN (IFN-λ) [61]. The IFN cascade mechanism is initiated when ligands (IFN type I/II/III) bind to their corresponding ligand receptor complex (interferon-alpha/beta receptor subunit 1 (IFNAR1) and interferon-alpha/beta receptor subunit 2 (IFNAR2) chains; interferon gamma receptor 1 (IFNGR1) and interferon gamma receptor 2 (IFNGR2) chains; and interleukin 10 receptor 2 (IL10R2) and interferon lambda receptor 1 (IFNLR1) chains, respectively) causing activation of receptor-associated Janus kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2) in the case of type I and III IFN; and Jak 1 and Jak 2 in the case of type II IFN-γ (Fig. 3). These kinases subsequently phosphorylate and activate the signal transducers and activators of transcription STAT1 and STAT2 (IFN type I and III) or the STAT1 dimer (IFN-γ) [62]. After activation, the STAT1 and STAT2 proteins heterodimerize and combine with IFN regulatory factor 9 (IRF9) to form a complex called IFN-stimulated gene factor 3 (ISGF-3). This complex then moves into the nucleus and identifies and binds to IFN-stimulated response elements (ISREs) in the target gene [62]. When it comes to IFN-γ, the STAT1 dimer, which is also referred to as the gamma interferon activation factor (GAF) complex, moves into the nucleus and binds with the gamma interferon activation site (GAS) enhancer elements of the target gene [63]. The ISGF-3 and GAF complex binding with their respective target gene sites results in the transcription of the IFN-stimulated gene (ISG) products, which act as antiviral agents [63].
It is important to note that the F and HN proteins are considered the primary determinants of NDV virulence, while information regarding the other proteins is limited [64]. Despite successfully entering the host body through binding and fusion, NDV must avoid the interferon (IFN) system, the host's first line of defense against viral infections. Researchers have used reverse genetics technology to learn how the virus evades the IFN-governed antiviral response, discovering that the V protein plays a critical role [14, 65]. Recombinant NDV lacking the V protein exhibited reduced growth in in vitro assays and embryonated chicken eggs, demonstrating the protein's importance in antagonizing the host's innate response [14]. The V protein targets phopho-STAT1, a protein that JAKs phosphorylate when stimulated by IFN-α, degrading it and reducing its accumulation, allowing NDV to evade the IFN system [66].
6. MECHANISM OF NDV IN TARGETING CANCER
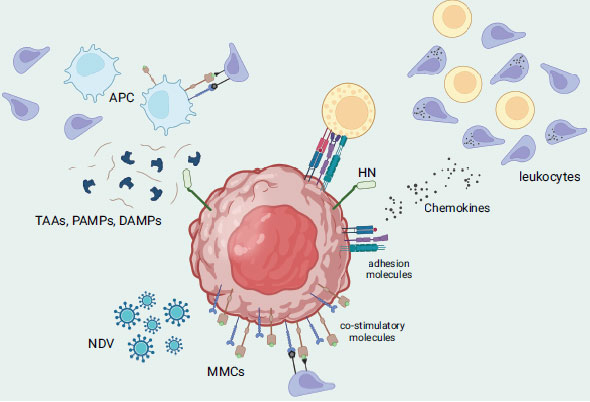
From laboratory studies to studies conducted in living organisms, all strains of NDV (velogenic, mesogenic, and lentogenic) have demonstrated significant potential in fighting multiple types of cancer. This is due to the defect in the interferon cascade found in tumor cells, which allows the NDV virus to successfully replicate within the cell before initiating its killing mechanism [67]. NDV's cancer-fighting activity can be triggered via direct or indirect mechanisms. The direct approach involves direct tumor-selective oncolysis, which includes the formation of multinucleated syncytia, activation of extrinsic and intrinsic apoptosis pathways, activation of the ER stress pathway, and involvement of the mitogen-activated protein kinase (MAPK) cascade [68, 69]. Fusogenic oncolytic viruses like NDV would express its F protein on the surface of the infected cells causing in the clumping/fusion of cells with their neighbours, thus forming large multi-nucleated cell bodies or syncytia, thereby rapidly activating apoptotic pathways [70, 71]. This would improve the viral spread and bystander killing of the oncolytic NDV tumor-killing machinery [72]. Other study underlines that the presence of both HN and F proteins is critical to have an efficient formation of stable syncytia whereas different ratios of F to HN result in varying outcomes of syncytia formation during NDV infection/treatment. On a related note, mutation in one of those proteins results in hyperfusogenic phenotypes (i.e., higher oncolytic potential). For example, the substitution of lysine with arginine led to the development of a mutated NDV that can facilitate cell-to-cell fusion even in the absence of HN [73]. Although the outcome of this mutation would be less efficient, it renders the use of mutated NDV for a more heterogenous tumor population (i.e., less specificity of F protein binding) [71]. Other than that, the presence of NDV in tumor cells may trigger the secretion of cytokines, including IFN-α, IFN-β, and TNF-α. These cytokines can activate the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) signaling pathway, which then stimulates the exogenous apoptotic pathway. On the other hand, previous research revealed that NDV's ability to induce apoptosis (i.e., intrinsic apoptotic pathway) may not depend on its replication or protein synthesis as NDV-infected cells can initially cause the activation of mitochondrial transition pores, followed by the activation of caspase-8 and subsequently, the expression of the viral NP gene [68]. Other than that, the indirect approach involves the innate and adaptive immune systems, where the infected cells release tumor-associated antigens and danger signals, leading to the activation of antigen-presenting cells and recruitment of immune cells such as T-cells, DCs, and NK cells (Fig. 4) [74, 75]. Several factors contribute to the increased cell surface adhesiveness of lymphocytes to tumor cells that are infected with NDV. These include the presence of viral receptor-binding glycoprotein HN, up-regulation of adhesion molecules such as intercellular adhesion molecule 1 (ICAM-1) and lymphocyte function-associated antigen 3 (LFA-3), and an increase in the secretion of pro-inflammatory cytokines and chemokines such as RANTES and IP-10. Additionally, NDV stimulates the up-regulation of major histocompatibility complex (MHC) and co-stimulatory molecules on tumor cells, allowing them to act as antigen-presenting cells and be recognized by tumor-specific lymphocytes of the adaptive immune system [76]. Besides the tumor cells, NDV can replicate itself in non-dividing tumour cells such as X-ray-irradiated tumour vaccine cells. Interestingly, NDV can target tumor stem cells or inactive tumor cells that may remain unaffected by chemotherapy or radiation therapy since viral replication in the cytoplasm does not rely on cell proliferation. Other than that, NDV is capable of multiplying in tumor cells that are resistant to apoptosis, as well as in cancer cells that lack oxygen (hypoxic) or are resistant to interferon [77].
7. PRE-CLINICAL STUDIES OF PARENTAL/WILD-TYPE NDV AGAINST CANCER
The use of the Newcastle disease virus (NDV) as an oncolytic agent for cancer therapy is still in the early stages of clinical development, and there is ongoing research to evaluate its safety and efficacy. However, preclinical studies have shown promising results in a variety of cancer types, including breast cancer, lung cancer, pancreatic cancer, and glioblastoma [78]. In these studies, NDV of various strains has been shown to selectively target and kill cancer cells while sparing healthy cells, induce apoptosis, stimulate the immune system, and inhibit angiogenesis [78]. For instance, in the early use of NDV, Cassel and his colleagues administered the 73T strain of NDV directly into the cervical cancer tumor area of patients for the first time whereby the findings indicated significant tumor elimination and a decrease in the metastasis of lymph nodes in the supraclavicular region, which served as preliminary evidence of the safety and potential of NDV as an anti-tumor agent [79]. Following this, Cassel suggested using autologous or allogeneic viral oncolysate (VOL) as a vaccination strategy. In the adjuvant immunotherapy of stage II malignant melanoma after surgery, NDV viral oncolysate was employed. After monitoring 83 patients for a decade, over 60% of them were found to have survived without experiencing a recurrence [80]. In another study, immunosuppressed severe combined immunodeficiency (SCID) in mice, which had been implanted with human neuroblastoma or fibrosarcoma, exhibited positive responses to a solitary injection of the NDV 73T virus strain directly into the tumor site whereby, they experienced either full or partial recovery [54]. On the other hand, a repeated administration of the mesogenic NDV MTH-68/H virus strain through intravenous injection led to a partial response in a 14-year-old boy who had glioblastoma [81]. Other than that, between 1990 and 2008, Schirrmacher's team focused on developing an autologous tumor vaccine (ATV), which consisted of 10 million irradiated tumor cells that had been previously infected with the weakened NDV strain Ulster. They administered the vaccine to patients with eight different types of cancer, including breast, colon, rectal, kidney, head and neck, pancreatic, and glioblastoma multiforme. The findings indicated that the use of a high-quality ATV-NDV vaccine could significantly enhance the survival outlook for patients as the median progression-free survival (PFS) and median overall survival (OS) were two times higher in the control group and no severe adverse effects were reported [82]. In addition, PV701 strain, which was derived from 73T strain was examined in three phase I clinical trials for patients with advanced malignancies through intravenous administration which gave one complete, one partial, and seven minor responses out of the 79 patients [83-85]. Fourteen patients showed prolonged progression-free survival that lasted from 4 to over 30 months [85]. In a subsequent study involving 18 patients with different advanced cancers, a higher response rate was observed with a slower infusion rate but higher therapeutic dosing [83, 84]. Four major and two minor responses were demonstrated, and six patients survived for at least two years. Other than that, fourteen patients with recurrent glioblastoma were treated with the lentogenic NDV strain HUJ using an intravenous approach, resulting in a complete response in one patient. In general, studies that have evaluated the intravenous administration of NDV have reported that it is well-tolerated, with flu-like symptoms being the most commonly reported adverse event [86]. Inspired by the oncolytic NDV research developments in the United States and Europe, Malaysia NDV AF2240 strain has been converted from being a virus used for vaccine purposes into an oncolytic research tool. The virus was tested against several types of cancer cells, including MCF-7, SW620, SW480, HL60, HeLa, and MDA-MB-231 whereby it demonstrated varying degrees of oncolytic effectiveness on different cell lines, with MDA-MB-231 showing greater susceptibility to viral treatment compared to MCF-7 [87-89]. Furthermore, when examined using electron microscopy, the cancer cells that were infected displayed typical morphological features of apoptosis, such as cell shrinkage, nuclear fragmentation, chromatin condensation, blebbing of the cell membrane, and the formation of apoptotic bodies [87, 88]. Ch’ng and his colleague reported that the AF2240 strain was able to effectively eliminate renal carcinoma cells (RCC) cells under both normoxic and hypoxic conditions, indicating its capacity to induce oncolysis independent of oxygen levels [90]. It is noteworthy that the virus exhibited increased oncolytic efficacy in hypoxic cells. Furthermore, another study demonstrated that NDV infection resulted in the degradation of hypoxia-inducible factor 1-alpha (HIF-1α) in a manner that was not dependent on the presence of p53 [91].
8. CLONING AND DEVELOPMENT OF RECOMBINANT NDV (rNDV) IN TARGETING CANCER
Substantial progress has been made in the development of recombinant NDV (rNDV) since Peeters et al. accomplished the first successful cloning of cDNA in 1999. As such, the lentogenic strain LaSota was employed to produce rNDV via the reverse genetic approach, which involves the recovery of the virus from a DNA copy of the RNA genome [92]. The term “reverse genetic” was first used by Weissmann et al. in 1978 to describe this process [92]. Over time, diverse methods have been utilized to construct the complete genome of rNDV. These include sequential cloning based on natural restriction enzyme sites, ligation-independent cloning, and synthetic production. This step enables the integration of novel or foreign genes or alteration of the NDV gene. Several studies have demonstrated that the highest foreign protein expression and NDV replication efficiency is achieved when the foreign DNA fragment is inserted nearer to the 3' end of the NDV genome [93]. Nevertheless, some research has suggested that optimal expression of the target protein can be attained by inserting foreign DNA fragments at other gene junction sites. For example, Zhao et al. and Wu et al. found optimal expression at the P/M and HN/L gene junctions, respectively [94, 95]. With regards to the successful use of parental/wild-type NDV as oncolytic tumor vaccine or adjuvant therapy, it is imperative to further maximise their therapeutic efficacy with the aid of genetic engineering. Table 2 outlines the genetic modification techniques used to develop a recombinant Newcastle disease virus and their potential applications. For instance, Janke and his colleague observed that there is no impact on the fundamental aspects of NDV replication or its preference for targeting tumors as a result of arming the NDV with gene encoding granulocyte/macrophage colony-stimulating factor (GM-CSF) [96]. Moreover, the resulting gene product demonstrated both biological activity and stability. In an in vitro tumor neutralization assay, tumor vaccine cells that were infected with rec(GM-CSF) caused human peripheral blood mononuclear cells (PBMC) to elicit antitumor bystander effects with enhanced performance compared to the control. Additionally, the rec(GM-CSF) also increased the interferon-α production in PBMC. Other than that, in vivo experiments showed that the NDV Anhinga strain, which expressed interleukin-2 (IL-2), effectively suppressed the growth of hepatocellular carcinoma while the rNDV-IL2-TRAIL variant greatly boosted the activation of programmed cell death (apoptosis) in cancer cells [95, 97, 98]. Meanwhile, augmentation of interleukin-7 (IL-7) and interleukin-15 (IL-15) in NDV strain LX (LX/IL7/IL15) exhibited effective anti-tumor properties against melanoma cells in mice [99]. On the other hand, NDV-MIP3α effectively secretes macrophage inflammatory protein 3-alpha (MIP-3α), resulting in robust promotion of dendritic cells (DCs) maturation and activation in B16 and CT26 cells. Notably, the intra-tumoral injection of NDV-MIP3α led to substantial suppression of tumor growth as well as a significant reversion of tumor microenvironments in B16 and CT26 mice models [100]. The anti-tumor effect of lentogenic rClone30 NDV was significantly improved with the presence of suicide genes namely, cytosine deaminase/5-fluorocytosine (CD/5-FC). Administering of this recombinant NDV, rClone30-CD/5-FC to mice with tumors results in a significant inhibition and sustained reduction of the tumors, leading to improved survival rates among the treated mice [101]. Another study carried out by Meng et al. revealed that the introduction of rNDV-VEGF-Trap resulted in a remarkable 85.37% reduction in cell growth ratio and an 87.9% decrease in migration ratio in EA.hy926 cells and also caused a reduction of tumor volume and weight by over threefold in CT26-bearing mice [102]. Furthermore, there is a significant decrease in the number of vascular endothelial cells within the tumor tissues and phosphorylation levels of Akt, extracellular signal-regulated kinase 1/2 (ERK1/2), and STAT3 while increased expression levels of P53, Bcl-2 associated X (BAX), and cleaved caspase-3 upon treatment with rNDV-VEGF-Trap [102]. On the other hand, rFMW/GFP infection resulted in a significant increase in early and late apoptosis in the anaplastic thyroid cancer (ATC), THJ-16 T, and THJ-29 T cell lines, as well as increased processing of caspase-3 and cleavage of Poly (ADP-ribose) polymerase (PARP) in the ATC cells. Of note, ATC stands out as one of the most aggressive solid tumors, lacking any currently available effective treatments [103]. Meanwhile, Numpadit et al. constructed a recombinant NDV that would release IFN-γ and target melanoma cells while recombinant NDV Italien strain carrying cHAb18 gene (rNDV-18HL) generated by Wei et al. could inhibit the invasion and migration of hepatocellular carcinoma (HCC) cells through affinity binding of mouse-human chimeric HAb18 antibody towards tumor-associated antigen CD147 [104, 105].
| Genetic Modification Techniques | Description | Potential Applications |
|---|---|---|
| Reverse genetics | Recombinant NDV viruses are generated by cloning the full-length cDNA of the viral genome into a plasmid vector. The plasmid is then transfected into cells expressing the viral polymerase complex, which generates the recombinant virus. | Generation of vaccine candidates, the study of viral pathogenesis, and development of oncolytic viruses [106]. |
| Site-directed mutagenesis | Site-directed mutagenesis is a technique that allows for the introduction of specific mutations into the viral genome. This can be used to study the function of viral genes or regulatory elements, or to generate recombinant viruses with altered properties. | Study of viral pathogenesis, identification of virulence determinants, and generation of attenuated vaccine strains [107]. |
| Insertion of foreign genes | Foreign genes can be inserted into the NDV genome to create recombinant viruses expressing heterologous proteins or antigens. This can be achieved by using reverse genetics techniques or by co-transfecting cells with the NDV genome and a plasmid expressing the foreign gene. | Generation of vaccine candidates expressing foreign antigens, development of recombinant viruses for gene therapy or as vectors for expression of therapeutic proteins [108]. |
| Deletion mutagenesis | Specific regions of the NDV genome can be deleted by genetic engineering techniques, such as reverse genetics or CRISPR-Cas9 genome editing. | Study of viral pathogenesis, identification of essential viral genes, generation of attenuated vaccine strains [109]. |
| Codon optimization | Codons in the NDV genome can be optimized to increase expression of viral proteins or foreign antigens in infected cells. This can be achieved by modifying the nucleotide sequence of the genome to match the codon usage of the host cell. | Generation of vaccine candidates expressing foreign antigens, development of recombinant viruses for gene therapy or as vectors for expression of therapeutic proteins [110]. |
| RNA interference | RNA interference (RNAi) can be used to knockdown expression of viral genes by introducing small interfering RNA (siRNA) molecules targeting specific viral mRNAs. | Study of viral pathogenesis, identification of essential viral genes [111]. |
| Chimeric NDV | The genome of NDV can be manipulated to replace certain genes or gene segments with those from related avian paramyxoviruses. This can generate chimeric NDV strains with altered antigenicity, pathogenicity, or replication properties. | Generation of vaccine candidates with improved efficacy or broadened antigenic coverage, study of viral pathogenesis [112, 113]. |
| RNA editing | RNA editing is a technique that allows for the modification of the RNA sequence of a virus without altering its DNA. RNA editing has been used to modify NDV genomes to reduce their pathogenicity or increase their immunogenicity. | Generation of vaccine candidates with improved safety or efficacy [114]. |
9. CHALLENGES OF NDV VIROTHERAPY IN TRANSLATIONAL RESEARCH
Despite the ongoing advancements in treatment strategies such as surgical, radiotherapy, immune checkpoint inhibitors, and chemotherapy, cancers have developed multiple mechanisms to evade therapies and the immune system of the host. Oncolytic viruses like NDV (wild-type and recombinant) offer a valuable alternative for cancers that show resistance to current treatments. In recent years, oncolytic NDVs have garnered significant attention due to their capacity to specifically target and destroy cancer cells, along with their potential to augment the immune response against tumors (i.e. excellent tool for immune-based therapy) [115]. The growing interest in NDVs stems from the advantages they hold over conventional cancer treatments, as certain tumor types remain unresponsive to these treatment strategies and pose several potential limitations, such as drug resistance, cancer recurrence, and severe adverse effects [115, 116]. Multiple clinical trials have provided evidence of NDVs utilizing diverse lytic mechanisms to eliminate cancer cells that exhibit resistance to traditional and targeted therapies (i.e. through limiting cell metabolism and generating a series of immunological responses), which suggests that NDVs have the potential to surmount certain obstacles in cancer treatment. Moreover, by employing rational design and genetic engineering techniques, NDVs can be customized to suit an individual's specific tumor type and its driver mutations, where modifications can be made to viral genes or noncoding sequences, enabling the addition or removal of specific functions, while non-viral genes or non-coding regulatory elements can be introduced to enhance desirable properties [117]. Furthermore, the development of predictive tests could enable the identification of patients with particular molecular profiles that make them more likely to derive benefits from NDV treatment, thus paving the way for a personalized medicine approach [118]. While oncolytic viruses have been recognized as a promising approach for targeted cancer therapy, achieving both high efficacy and specificity with this strategy presents significant challenges. To date, there are only four oncolytic viruses approved namely, ECHO-7 (unmodified picornavirus), engineered adenovirus H101, T-VEC (genetically engineered herpes simplex virus type-1), and teserpaturev to treat melanoma, head and neck cancer, unresectable melanoma, and malignant gliomas, respectively [118]. Remarkably, none of them are NDVs, which suggests that it has quite a long way to go before the NDV virotherapy gets the approval and is in the market. Of note, numerous oncolytic viruses have halted their development due to either insufficient effectiveness or an unacceptable level of toxicity [118]. The process of developing OVs presents several challenges, which can contribute to the difficulties in obtaining approval. For example, numerous pre-clinical studies have demonstrated the detrimental effects of the host immune response, including decreased replication and earlier clearance of the viruses, as well as reduced anti-tumor effectiveness in immunocompetent subjects compared to immunocompromised subjects [119, 120]. Therefore, the advancement of oncolytic NDV treatment strategies should prioritize the exploration of methods to modulate the host immune system, whereby the goal is to reduce antiviral responses and viral clearance while simultaneously promoting immune reactions that facilitate the destruction of tumors [119]. As such, an approach to prevent rapid clearance of the virus involves the temporary suppression of early immune responses. For instance, a preclinical demonstration of this strategy involved the use of a recombinant VSV (vesicular stomatitis virus) engineered to express a chemokine-binding protein derived from herpesvirus-1 glycoprotein G [121]. By suppressing the host's antiviral inflammatory response, the oncolytic virus exhibited increased potency, leading to significantly prolonged survival in rats with multifocal hepatocellular carcinoma. Other than that, further exploration of NDV would focus on identifying the optimal delivery methods and dosages to achieve maximum efficacy. It is crucial to establish an optimal dosing schedule in order to take advantage of the timeframe between initial administration and the production of anti-NDV antibodies when utilizing isolated strains of NDV for their cytotoxic and immunostimulatory effects [122]. This approach would enhance the delivery of infectious particles to the tumor site. For instance, Ghrici et al. conducted a study revealing the activation of caspase-8 within 2 hours of infecting MCF7 cells with the AF2240 strain, which contrasts with the findings presented by Elankumaran et al. and Ravindra, where caspase-8 activation was observed only after 48 and 24 hours, respectively [123]. This temporal discrepancy in caspase-8 activation may not necessarily be attributed to variations in the sequence of external and internal apoptosis pathways but could potentially be linked to the impact of strain virulence on the rate of apoptosis [123]. The variation in the timing of the activation of the cell death program could hold significance in the suppression of tumor progression. Other than that, it is also feasible to aim to investigate whether specific genetic or molecular tumor profiles exhibit higher susceptibility to NDV-based immunotherapy, aligning with the principles of personalized therapy [124]. In addition, current data indicates that the administration of tumor NDV vaccines results in significant activation of the immune response in the surrounding areas (i.e. peripheral immune response) [122]. It is crucial to comprehend both the strength of this immune response in the surrounding regions and its effect within the tumor environment. Furthermore, it is essential to explore how this activated immune system influences any remaining tumor cells and its potential role in preventing tumor recurrence. On the other hand, the integration of genetic engineering techniques with computational approaches has the potential to improve the effectiveness of cancer treatment in clinical settings. Additionally, the best combination partners between NDV virotherapy and existing modalities/immunotherapies need to be explored to achieve broader application prospects, such as overcoming chemoresistance and increasing tumor cell sensitivity to radiation therapy. By combining therapeutic agents with a different mechanism of action, efficacy may be enhanced and yield more successful therapeutic outcomes [115]. It is also noteworthy that the excessive expression of foreign genes will affect the replication of NDV, which poses a challenge in developing a cancer therapeutic recombinant NDV [116]. Furthermore, achieving a clinical-grade virus preparation of NDV requires thorough purification processes [125].
CONCLUSION
NDV poses significant threats in poultry industries with multiple degrees of virulence among avian species. The biology of NDV widens our research opportunities in search for better vaccine development and diagnostic tools especially in cancer research. NDV demonstrates therapeutic compatibility for novel drugs i.e. oncolytic properties with high selectivity, as well as broad immunostimulatory activities, which promotes a new generation of biosafety agents. Moreover, the advent of genetic engineering allows the modification of NDV for better immunogenic potential of innate and adaptive immunity. The technology of reverse genetics utilizing NDV as vectors enables the incorporation of therapeutic genes. Although NDV portrays promising oncolytic potential, however, preclinical studies involving their optimum route of administration, genetic engineering tools, as well as their safety efficacy are still lacking. Thus, it is imperative to vigorously explore the remaining questions to move the field forward, promoting a translational approach towards clinical settings.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This study was funded by Universiti Malaysia Sabah, Awards/Grant number.: Skim Dana Nic (DN20089) and Skim Dana Khas (SDK0299-2020).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


