All published articles of this journal are available on ScienceDirect.
Heavy Metal Tolerance and Removal Efficiencies by Soil Bacterial Strains: Effects of Carbon and Nitrogen Sources
Abstract
Background:
Several human activities contribute to the release of heavy metals into the environment, which constitutes a threat to the environment and human health; thus, there is a need for remediation of these metals.
Methods:
The aim of this study was to evaluate the effects of carbon and nitrogen sources on tolerance to lead, nickel and cadmium by soil bacterial strains. The effects of carbon, nitrogen sources and carbon-nitrogen ratio on the bacteria strains were also explored. A total of ten bacterial species, which comprise Yersinia enterocolitica (1), Alcaligenes faecalis (4), Bacillus cereus (2), Enterobacter cloacae (1) and Bacillus subtilis (2), were identified. The screening was carried out in minimal media using different carbon sources (sodium acetate, glucose, sucrose and maltose), nitrogen sources (yeast extract, peptone, tryptone and potassium nitrate) and carbon/nitrogen (C/N) ratios (5:5, 5:4, 5:3 and 5:2). Based on tolerance index, the optimal carbon and nitrogen sources were observed to be sodium acetate and potassium nitrate, respectively, while the C/N ratio varied across the isolates.
Results & Discussion:
At the end of the study, the tolerance index observed for cadmium, lead, and nickel ranged from 0.44 to 0.55, from 0.48 to 2.27 and from 0.19 to 1.95, respectively. Moreover, removal percentages that ranged from 12%-35%, 56%-97% and 79%-90% were observed for cadmium, lead and nickel, respectively, in the presence of the bacterial species.
Conclusion:
The results showed the bacterial isolates' effectiveness in removing these heavy metals from the environment.
1. INTRODUCTION
Modern activities such as urbanization and industrialization have greatly contributed to the release of heavy metals into the environment, resulting in their assemblage and distribution within the environment [1]. The release of these metals into the environment is also a grave risk to public health. Although heavy metal sources in the environment can also be from natural sources (weathering of rocks, volcanic eruptions, etc.) or from a man-made source, which may be from anthropogenic activities of man such as agricultural use of pesticides, fertilizers and herbicides, mining, paints, sewage sludge, solid waste disposal, burning of fossil fuels, etc. [2].
These anthropogenic sources highly contribute to introducing heavy metals into the environment since most of these activities generate waste that contains heavy metals that harm the environment and human well-being. They are incessant in the environment and can pollute and accumulate in the food web, thus causing health issues because of their toxic nature [3]. Soil contaminated with heavy metals may be dangerous to animals, humans, and the ecosystem via exposure routes such as direct contact or ingesting contaminated soil or contaminated groundwater [4]. Heavy metals lower food standards in terms of safety and marketability due to inhibited plant growth; they also reduce the number of available lands for agricultural purposes, which may result in problems of food insecurity [4].
Several conventional methods and processes, such as solvent extraction, ion exchange, oxidation-reduction, filtration, and chemical precipitation, have been developed to enable the removal/elimination of these metals from the environment, eliminating the risk to public health and improving environmental sustainability [5]. However, these conventional methods have their shortcomings as they are considered less effective, costly and lacking in removing low concentrations of heavy metals [6]. Some of these methods generate toxic by-products, such as sludge. Recent studies using microbial remediation (which involves the use of bacteria, algae, fungi, etc.) have proved to be very efficient in removing these heavy metals from the environment. This approach is considered environmentally friendly, of low cost, and effective in mitigating heavy metals to acceptable levels in the environment.
Microbial remediation is a bioremediation technique that uses microorganisms to remove heavy metals from the soil through oxidation, absorption, and precipitation processes. They possess metabolic abilities that enable them to use toxic compounds through metabolism, respiration and fermentation [7]. Several studies have used microorganisms such as Flavobacterium, Enterobacter, Bacillus, Pseudomonas, Micrococcus,etc., to treat heavy metal-contaminated soil. For instance, Alcaligenes faecalis and Bacillus sp. have been used for remediating heavy metal-contaminated soil [8]. A study observed 70% and 75% removal efficiencies in cadmium reduction by Pseudomonas aeruginosa and Alcaligenes faecalis [9]. The removal of heavy metals by bacterial strains can be influenced by the presence of carbon and nitrogen sources. Bacteria derive energy for growth and metabolic activities from carbon sources, while nitrogen sources provide bacteria with the production of enzymes and protein synthesis [8]. The type and concentration of carbon and nitrogen sources present in the medium can also enhance the heavy metal removal efficiencies of bacteria strains. Therefore, it is essential to select an optimal carbon and nitrogen source in order to enhance the removal efficiencies of heavy metals by bacterial strains. Therefore, This study aimed to isolate and screen indigenous soil bacteria for tolerance and removal of selected heavy metals in liquid media. The effects of carbon, nitrogen sources and carbon-nitrogen ratio on the bacteria strains were also explored.
2. MATERIALS AND METHODS
2.1. Bacterial Isolation and Screening for Heavy Metal Tolerance and Removal
A total of 65 bacterial strains were isolated from soil samples within Afe Babalola University environs. Bacterial isolation was carried out using the standard pour-plating technique. Following isolation, representative colonies were streaked on nutrient agar plates and incubated at 37 °C for 24h to obtain pure cultures. The pure cultures were stored on nutrient agar slants at 4 °C ±2 °C until needed.
For preliminary screening for heavy metal tolerance and removal, lead, nickel, and cadmium were used in this study. Nutrient broth, supplemented with different concentrations (60 ppm, 90 ppm, 120 ppm, 150 ppm, 180 ppm, 210 ppm) of the respective metal salts (lead nitrate (Pb (NO3)2, Cadmium chloride hemihydrate (CdCl2. 21/2 H2O) and Nickel sulfate (NiSO4) was prepared and sterilized using an autoclave. After sterilization, 10mL of the respective metal concentrations were dispensed in 20mL capacity sterile universal bottles, inoculated with 0.5 mL of overnight grown pure cultures of the respective isolates (in duplicates), and incubated at 35 °C ±2 °C for 48h. At the expiration of incubation, the growth rate was read at 700nm using a UV/VIS Spectrophotometer and the residual metal concentration was determined using Atomic Absorption Spectrometry (AAS). The metal tolerance index (TI) and percent removal were estimated for each metal in the presence of the respective isolates.
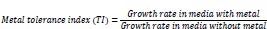 |
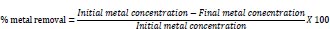 |
Only 10 isolates that showed significant tolerance and removal of metals were identified and used for optimization studies.
2.2. Optimization for Tolerance and Removal Studies
Three parameters: carbon source, nitrogen source and carbon/nitrogen ratio (C/N) were optimized for in this study. The carbon sources used for investigation were glucose, maltose, sodium acetate and sucrose, while the nitrogen sources were potassium nitrate (KNO3), peptone, yeast extract and tryptone. The C/N ratios used were 5:5, 5:4, 5:3 and 5:2.
Optimization studies were carried out separately (for each of the carbon and nitrogen sources using the different C/N ratios stated above) in minimal media composed of carbon source (5 g/L), nitrogen source (2 g/L), magnesium sulphate (0.5 g/L), water (1 L) and heavy metal (150 mg/L). Neutral pH was used in this study. Following media preparation and sterilization, the respective isolates were used for inoculation and incubated at 35 °C ±2 °C for 96 h. At the expiration of incubation, the growth rate of the isolates and the residual concentration of metals in both the inoculated and uninoculated setups were determined to estimate the metal tolerance index and percentage of metal removal.
2.3. Characterization of Isolates
The isolates were characterized using the Sanger sequencing method. Isolate sequences were deposited in the National Centre for Biotechnology Information (NCBI) database, and Accession numbers from OQ383311 to OQ383320 were obtained.
2.4. Statistical Analysis
All statistical analyses were carried out using the SPSS statistical software (version 23.0). Comparison of means was determined using the One-Way Analysis of Variance (ANOVA) test, while multiple comparison was determined using the Tukey Multiple Range test. All analyses were carried out at a 95% confidence interval.
| Isolates Code | Isolates | Max Score | Total Score | Query Cover | % Identity | Accession |
|---|---|---|---|---|---|---|
| A | Yersinia enterocolitica | 2545 | 21787 | 99% | 98.35% | OQ383311 |
| B | Alcaligenes faecalis | 2287 | 2287 | 99% | 95.31% | OQ383312 |
| C | Bacillus cereus | 2263 | 2263 | 99% | 98.01% | OQ383313 |
| D | Alcaligenes faecalis | 2619 | 2619 | 99% | 99.86% | OQ383314 |
| E | Bacillus subtilis | 2143 | 2143 | 100% | 97.99% | OQ383315 |
| F | Enterobacter cloacae | 2366 | 18878 | 98% | 96.64% | OQ383316 |
| G | Bacillus cereus | 2468 | 2468 | 99% | 99.49% | OQ383317 |
| H | Alcaligenes faecalis | 1668 | 1668 | 98% | 99.56% | OQ383318 |
| I | Alcaligenes faecalis | 2459 | 7377 | 99% | 97.82% | OQ383319 |
| J | Bacillus subtilis | 2586 | 2586 | 100% | 98.57% | OQ383320 |
3. RESULTS
3.1. Test Isolates
A total of ten bacterial species, which comprise Yersinia enterocolitica (1), Alcaligenes faecalis (4), Bacillus cereus (2), Enterobacter cloacae (1) and Bacillus subtilis (2), were identified (Table 1).
3.2. Effect of External Carbon Sources
In the presence of the respective carbon sources, the significantly highest tolerance index was observed for nickel in media that contained sodium acetate as its carbon source (p≤0.05); this observation was irrespective of the bacterial species. Across the respective isolates, the tolerance indices for nickel in the media that contained sodium acetate ranged between 0.50 and 1.42, observed in the presence of isolates F and A, respectively. In the case of the tolerance index for lead, significantly highest (p≤0.05) values were recorded in media that contained sodium acetate in the presence of most of the isolates, with the exception of isolates E and F, where the significantly highest tolerance values were recorded in media that contained maltose and glucose, respectively. In the case of cadmium, a significantly higher tolerance index was also observed in media that contained sodium acetate. This observation was also irrespective of the test isolates, except for isolate F, where the highest tolerance was observed in the media that contained glucose (Table 2).
With respect to removal of heavy metals, % removal ranged from 51.43 (Isolate C in media that contained glucose) to 88.43 (Isolate F in media that contained maltose), from 38.03 (Isolate J in media with glucose) to 88.60 (Isolate G in media with sucrose) and from 65.93 (Isolate E in media with glucose) to 92.03 (Isolate H in media with sucrose) were observed in presence of the test bacterial species for lead, cadmium and nickel, respectively. Generally, significantly (p≤0.05), the highest removal of lead was observed in media with sucrose (Isolates B, C, D, G and J), maltose (Isolates A, E, F and I) and glucose (Isolate H). Cadmium removal was, however, observed to be the highest in media containing sucrose (for Isolates A, C, D, G, H and J) and maltose (for Isolates B, E, F and I). In the case of nickel removal, significantly highest values were observed in media that contained sucrose (for Isolates A, B, E, F, G, H and J), acetate (for Isolates C and D) and maltose (for Isolates I) (Table 3).
| Bacterial Strains | Carbon Sources | |||
|---|---|---|---|---|
| Glucose | Maltose | Acetate | Sucrose | |
| Nickel | ||||
| Yersinia enterocolitica (OQ383311) | 0.75±0.01a | 0.31±0.00b | 1.42±0.01c | 0.34±0.01d |
| Alcaligenes faecalis (OQ383312) | 0.35±0.00a | 0.42±0.00b | 0.89±0.01c | 0.47±0.01d |
| Bacillus cereus (OQ383313) | 0.64±0.01a | 0.49±0.01b | 1.18±0.04c | 0.09±0.00d |
| Alcaligenes faecalis (OQ383314) | 0.23±0.00a | 0.27±0.00b | 0.58±0.01c | 0.23±0.00a |
| Bacillus subtilis (OQ383315) | 0.64±0.01a | 0.49±0.01b | 1.18±0.04c | 0.09±0.00d |
| Enterobacter cloacae (OQ383316) | 0.58±0.00a | 0.27±0.00b | 0.50±0.00c | 0.26±0.00d |
| Bacillus cereus (OQ383317) | 0.42±0.00a | 0.43±0.00b | 0.79±0.00c | 0.39±0.00d |
| Alcaligenes faecalis (OQ383318) | 0.34±0.00a | 0.40±0.00b | 0.74±0.01c | 0.35±0.00d |
| Alcaligenes faecalis (OQ383319) | 0.47±0.00a | 0.53±0.00b | 0.74±0.00c | 0.41±0.00d |
| Bacillus subtilis (OQ383320) | 0.28±0.00a | 0.53±0.01b | 0.70±0.00c | 0.50±0.01d |
| Lead | ||||
| Yersinia enterocolitica (OQ383311) | 0.86±0.01a | 0.55±0.00b | 2.01±0.02c | 0.57±0.00d |
| Alcaligenes faecalis (OQ383312) | 0.28±0.00a | 0.94±0.00b | 1.02±0.01c | 0.79±0.00d |
| Bacillus cereus (OQ383313) | 0.52±0.00a | 0.49±0.01a | 1.18±0.04b | 0.09±0.00c |
| Alcaligenes faecalis (OQ383314) | 0.25±0.00a | 0.53±0.01b | 0.78±0.01c | 0.28±0.00d |
| Bacillus subtilis (OQ383315) | 0.76±0.01a | 1.00±0.01b | 0.69±0.00c | 0.51±0.00d |
| Enterobacter cloacae (OQ383316) | 0.71±0.01a | 0.64±0.01b | 0.67±0.00c | 0.55±0.00d |
| Bacillus cereus (OQ383317) | 0.54±0.00a | 0.56±0.01b | 0.83±0.01c | 0.93±0.01d |
| Alcaligenes faecalis (OQ383318) | 0.43±0.00a | 0.42±0.01b | 0.75±0.00c | 0.27±0.00d |
| Alcaligenes faecalis (OQ383319) | 0.89±0.00a | 0.68±0.00b | 1.01±0.00c | 0.52±0.01d |
| Bacillus subtilis (OQ383320) | 0.62±0.01a | 0.61±0.01a | 0.82±0.01c | 0.57±0.00d |
| Cadmium | ||||
| Yersinia enterocolitica (OQ383311) | 0.26±0.01a | 0.08±0.00b | 0.34±0.01c | 0.12±0.00d |
| Alcaligenes faecalis (OQ383312) | 0.18±0.00a | 0.19±0.00b | 0.32±0.00c | 0.21±0.00d |
| Bacillus cereus (OQ383313) | 0.41±0.00a | 0.20±0.00b | 0.43±0.00c | 0.08±0.00d |
| Alcaligenes faecalis (OQ383314) | 0.10±0.00a | 0.10±0.00a | 0.23±0.00b | 0.08±0.00c |
| Bacillus subtilis (OQ383315) | 0.39±0.01a | 0.31±0.00b | 0.44±0.00c | 0.21±0.00d |
| Enterobacter cloacae (OQ383316) | 0.29±0.01a | 0.11±0.00b | 0.18±0.00c | 0.13±0.00d |
| Bacillus cereus (OQ383317) | 0.28±0.00a | 0.25±0.00b | 0.46±0.00c | 0.25±0.00d |
| Alcaligenes faecalis (OQ383318) | 0.23±0.00a | 0.19±0.00b | 0.47±0.00c | 0.20±0.00d |
| Alcaligenes faecalis (OQ383319) | 0.28±0.00a | 0.24±0.00b | 0.30±0.00c | 0.15±0.00d |
| Bacillus subtilis (OQ383320) | 0.29±0.00a | 0.29±0.01a | 0.40±0.00b | 0.24±0.00c |
| Bacterial Strains | Carbon Sources | |||
|---|---|---|---|---|
| Acetate | Glucose | Sucrose | Maltose | |
| Nickel | ||||
| Yersinia enterocolitica (OQ383311) | 76.10±0.27a | 66.60±0.08b | 86.60±0.23c | 81.57±0.04d |
| Alcaligenes faecalis (OQ383312) | 80.67±0.00a | 83.50±0.04b | 88.10±0.12c | 78.93±0.23d |
| Bacillus cereus (OQ383313) | 88.27±0.08a | 72.23±0.27b | 79.13±0.23c | 75.40±0.08d |
| Alcaligenes faecalis (OQ383314) | 82.87±0.54a | 69.60±0.31b | 82.07±0.23c | 79.13±0.08d |
| Bacillus subtilis (OQ383315) | 75.40±0.23a | 65.93±0.15b | 83.67±0.38c | 80.57±0.12d |
| Enterobacter cloacae (OQ383316) | 87.17±0.19a | 81.73±0.31b | 90.87±0.15c | 74.63±0.19d |
| Bacillus cereus (OQ383317) | 79.50±0.19a | 72.27±0.31b | 81.83±0.04c | 73.77±0.12d |
| Alcaligenes faecalis (OQ383318) | 84.80±0.15a | 84.97±0.04a | 92.03±0.12b | 82.43±0.27c |
| Alcaligenes faecalis (OQ383319) | 81.90±0.04a | 72.43±0.27b | 79.47±0.23c | 87.23±0.12d |
| Bacillus subtilis (OQ383320) | 79.10±0.27a | 69.43±0.27b | 81.83±0.19c | 75.23±0.12d |
| Lead | ||||
| Yersinia enterocolitica (OQ383311) | 61.03±0.19a | 58.53±0.62b | 73.27±0.31c | 87.73±0.31d |
| Alcaligenes faecalis (OQ383312) | 57.37±0.04a | 73.73±0.31b | 79.27±0.08c | 65.17±0.42d |
| Bacillus cereus (OQ383313) | 65.70±0.27a | 51.43±0.27b | 80.47±0.23c | 79.33±0.15d |
| Alcaligenes faecalis (OQ383314) | 58.27±0.31a | 61.60±0.31b | 71.67±0.23c | 61.80±0.23b |
| Bacillus subtilis (OQ383315) | 67.20±0.15a | 67.93±0.15b | 61.23±0.12c | 88.43±0.12d |
| Enterobacter cloacae (OQ383316) | 55.80±0.23a | 64.63±0.19b | 68.60±0.23c | 77.63±0.27d |
| Bacillus cereus (OQ383317) | 76.37±0.04a | 67.50±0.35b | 81.00±0.38c | 66.40±0.15d |
| Alcaligenes faecalis (OQ383318) | 58.57±0.12a | 82.23±0.04b | 70.83±0.19c | 75.20±0.15d |
| Alcaligenes faecalis (OQ383319) | 64.77±0.12a | 70.27±0.31b | 61.63±0.27c | 83.13±0.23d |
| Bacillus subtilis (OQ383320) | 66.60±0.08a | 58.83±0.12b | 76.33±0.38c | 71.57±0.19d |
| Cadmium | ||||
| Yersinia enterocolitica (OQ383311) | 79.13±0.15a | 48.33±0.23b | 87.30±0.04c | 76.90±0.27d |
| Alcaligenes faecalis (OQ383312) | 61.63±0.27a | 43.60±0.31b | 76.33±0.38c | 77.97±0.12d |
| Bacillus cereus (OQ383313) | 75.83±0.27a | 64.93±0.31b | 81.70±0.35c | 74.90±0.12d |
| Alcaligenes faecalis (OQ383314) | 73.87±0.15a | 72.53±0.23b | 87.30±0.19c | 56.90±0.27d |
| Bacillus subtilis (OQ383315) | 66.23±0.27a | 66.23±0.27a | 71.80±0.54b | 72.43±0.19c |
| Enterobacter cloacae (OQ383316) | 81.27±0.15a | 61.80±0.23b | 77.77±0.12c | 59.17±0.19d |
| Bacillus cereus (OQ383317) | 70.17±0.19a | 80.30±0.19b | 88.60±0.23c | 64.63±0.19d |
| Alcaligenes faecalis (OQ383318) | 59.00±0.38a | 68.60±0.08b | 77.37±0.12c | 51.33±0.15d |
| Alcaligenes faecalis (OQ383319) | 81.40±0.15a | 59.17±0.27b | 80.33±0.15c | 65.67±0.38d |
| Bacillus subtilis (OQ383320) | 58.90±0.27a | 38.03±0.19b | 65.27±0.69c | 59.87±0.15d |
3.3. Effect of External Nitrogen Sources.
In the different nitrogen sources used in this experiment, the highest tolerance index observed in the lead was the media containing potassium nitrate, which ranged between 0.19 and 1.71, observed in bacterial species B and A. For cadmium, the significantly highest (p≤0.05) values were observed in media containing potassium nitrate (0.68) in the presence of isolate G. In the case of nickel, the lowest and highest tolerance index values of 0.17 and 2.58 were observed in media containing peptone and potassium nitrate, respectively (Table 4).
| Bacterial Strains | Nitrogen Sources | |||
|---|---|---|---|---|
| KNO3 | Peptone | Yeast Extract | Tryptone | |
| Nickel | ||||
| Yersinia enterocolitica (OQ383311) | 2.58±0.02a | 0.30±0.00b | 0.36±0.00c | 0.47±0.01d |
| Alcaligenes faecalis (OQ383312) | 1.19±0.24a | 0.17±0.00b | 0.70±0.00c | 0.29±0.00b |
| Bacillus cereus (OQ383313) | 1.38±0.01a | 0.34±0.00b | 0.66±0.00c | 0.33±0.00b |
| Alcaligenes faecalis (OQ383314) | 1.39±0.02a | 0.29±0.00b | 0.74±0.00c | 0.33±0.01d |
| Bacillus subtilis (OQ383315) | 1.08±0.00a | 0.26±0.01b | 0.72±0.01c | 0.45±0.01d |
| Enterobacter cloacae (OQ383316) | 1.77±0.01a | 0.41±0.00b | 0.77±0.00c | 0.33±0.00d |
| Bacillus cereus (OQ383317) | 1.37±0.01a | 0.37±0.01b | 0.58±0.01c | 0.27±0.00d |
| Alcaligenes faecalis (OQ383318) | 1.21±0.00a | 0.25±0.00b | 0.60±0.00c | 0.22±0.01d |
| Alcaligenes faecalis (OQ383319) | 1.96±0.02a | 0.40±0.00b | 0.62±0.00c | 0.34±0.01d |
| Bacillus subtilis (OQ383320) | 1.12±0.03a | 0.55±0.00b | 0.61±0.00c | 0.42±0.00d |
| Lead | ||||
| Yersinia enterocolitica (OQ383311) | 1.71±0.01a | 0.75±0.01b | 0.64±0.01c | 1.40±0.02d |
| Alcaligenes faecalis (OQ383312) | 0.19±0.05a | 0.58±0.00b | 0.43±0.00c | 0.88±0.00d |
| Bacillus cereus (OQ383313) | 0.90±0.02a | 0.79±0.00b | 0.54±0.01c | 1.00±0.01d |
| Alcaligenes faecalis (OQ383314) | 1.04±0.01a | 0.60±0.00b | 0.50±0.01c | 0.63±0.01d |
| Bacillus subtilis (OQ383315) | 1.00±0.00a | 1.02±0.00b | 0.47±0.01c | 0.72±0.01d |
| Enterobacter cloacae (OQ383316) | 1.25±0.01a | 0.79±0.00b | 0.42±0.00c | 0.63±0.00d |
| Bacillus cereus (OQ383317) | 0.89±0.00a | 0.96±0.00b | 0.54±0.00c | 1.22±0.00d |
| Alcaligenes faecalis (OQ383318) | 0.91±0.01a | 0.68±0.00b | 0.74±0.00c | 0.77±0.00d |
| Alcaligenes faecalis (OQ383319) | 1.36±0.01a | 0.92±0.00b | 0.54±0.00c | 0.67±0.01d |
| Bacillus subtilis (OQ383320) | 0.94±0.02a | 1.64±0.01b | 0.69±0.01c | 0.59±0.00d |
| Cadmium | ||||
| Yersinia enterocolitica (OQ383311) | 0.38±0.00a | 0.46±0.00b | 0.10±0.00c | 0.06±0.01d |
| Alcaligenes faecalis (OQ383312) | 0.16±0.04a | 0.37±0.00b | 0.51±0.00c | 0.08±0.00d |
| Bacillus cereus (OQ383313) | 0.31±0.02a | 0.44±0.00b | 0.21±0.00c | 0.09±0.00d |
| Alcaligenes faecalis (OQ383314) | 0.23±0.02a | 0.55±0.00b | 0.15±0.00c | 0.05±0.00d |
| Bacillus subtilis (OQ383315) | 0.23±0.00a | 0.44±0.00b | 0.33±0.01c | 0.12±0.01d |
| Enterobacter cloacae (OQ383316) | 0.25±0.01 | 0.31±0.00b | 0.19±0.01c | 0.04±0.00d |
| Bacillus cereus (OQ383317) | 0.68±0.00a | 0.30±0.01b | 0.36±0.00c | 0.06±0.00d |
| Alcaligenes faecalis (OQ383318) | 0.55±0.01a | 0.29±0.01b | 0.45±0.01c | 0.09±0.00d |
| Alcaligenes faecalis (OQ383319) | 0.60±0.01a | 0.16±0.00b | 0.60±0.01a | 0.06±0.00c |
| Bacillus subtilis (OQ383320) | 0.42±0.01a | 0.18±0.00b | 0.10±0.01c | 0.12±0.00d |
As regards the removal of heavy metals by bacterial species, the removal percentage ranged between 49.43 (isolate G in media containing peptone), and 77.63 (isolate E in media containing potassium nitrate), 34.77 (isolate H in media containing tryptone) and 83.17 (isolate A in media containing yeast extract), 51.43 (isolate C in media containing peptone) and 90.20 (isolate F in media containing yeast extract) in lead, cadmium and nickel correspondingly. Significantly (p≤0.05), the highest removal of lead was observed in media with potassium nitrate (for isolates A, B, D, E, F, H, and J), peptone (isolate C), tryptone (isolates G and J). Moreover, the removal of cadmium was observed to be significantly highest in media with yeast extract in all test isolates, excluding isolate E, which was in tryptone. For removal in nickel, the significantly highest values were observed in media containing yeast extract for all test isolates (Table 5).
| Bacterial Strains | Nitrogen Sources | |||
|---|---|---|---|---|
| KNO3 | Peptone | Yeast Extract | Tryptone | |
| Nickel | ||||
| Yersinia enterocolitica (OQ383311) | 58.83±0.19a | 86.30±0.04b | 59.57±0.50c | 67.67±0.23d |
| Alcaligenes faecalis (OQ383312) | 72.23±0.27a | 83.33±0.15b | 64.23±0.27c | 74.47±0.54d |
| Bacillus cereus (OQ383313) | 59.83±0.19a | 85.70±0.27b | 51.43±0.27c | 74.03±0.42d |
| Alcaligenes faecalis (OQ383314) | 63.23±0.65a | 86.17±0.19b | 59.50±0.58c | 72.50±0.19d |
| Bacillus subtilis (OQ383315) | 71.70±0.19a | 83.07±0.46b | 61.93±0.08c | 74.47±0.23d |
| Enterobacter cloacae (OQ383316) | 65.67±0.23a | 90.20±0.23b | 69.93±0.31c | 83.33±0.00d |
| Bacillus cereus (OQ383317) | 68.30±0.27a | 86.33±0.23b | 57.73±0.15c | 79.07±0.15d |
| Alcaligenes faecalis (OQ383318) | 59.00±0.38a | 88.30±0.12b | 67.63±0.27c | 73.87±0.15d |
| Alcaligenes faecalis (OQ383319) | 63.93±0.23a | 85.53±0.15b | 65.30±0.27c | 72.33±0.15d |
| Bacillus subtilis (OQ383320) | 59.40±0.31a | 83.63±0.27b | 59.07±0.31a | 73.20±0.15c |
| Lead | ||||
| Yersinia enterocolitica (OQ383311) | 75.73±0.15a | 66.90±0.19b | 70.47±0.23c | 65.63±0.27d |
| Alcaligenes faecalis (OQ383312) | 74.33±0.38a | 68.17±0.12b | 60.60±0.31c | 67.33±0.38d |
| Bacillus cereus (OQ383313) | 65.57±0.27a | 67.63±0.35b | 69.67±0.38c | 61.90±0.19d |
| Alcaligenes faecalis (OQ383314) | 76.43±0.04a | 65.87±0.38b | 57.97±0.19c | 75.77±0.27d |
| Bacillus subtilis (OQ383315) | 77.63±0.27a | 66.03±0.19b | 58.93±0.23c | 64.63±0.27d |
| Enterobacter cloacae (OQ383316) | 71.43±0.27a | 67.40±0.31b | 52.93±0.31c | 59.20±0.15d |
| Bacillus cereus (OQ383317) | 65.37±0.19a | 67.73±0.46b | 49.43±0.42c | 68.23±0.19b |
| Alcaligenes faecalis (OQ383318) | 72.40±0.31a | 61.90±0.12b | 60.17±0.04c | 61.27±0.08d |
| Alcaligenes faecalis (OQ383319) | 75.40±0.23a | 61.00±0.23b | 65.50±0.19c | 75.13±0.23a |
| Bacillus subtilis (OQ383320) | 66.53±0.15a | 65.37±0.42b | 51.53±0.23c | 67.00±0.23d |
| Cadmium | ||||
| Yersinia enterocolitica (OQ383311) | 51.90±0.65a | 83.17±0.58b | 58.47±0.54c | 73.03±0.19d |
| Alcaligenes faecalis (OQ383312) | 64.63±0.19a | 66.57±0.12b | 50.93±0.31c | 61.60±0.23d |
| Bacillus cereus (OQ383313) | 59.60±0.23a | 81.80±0.23b | 59.43±0.27c | 64.17±0.19d |
| Alcaligenes faecalis (OQ383314) | 68.27±0.31a | 78.67±0.23b | 60.33±0.23c | 72.43±0.12d |
| Bacillus subtilis (OQ383315) | 57.87±0.15a | 61.17±4.04bc | 59.27±0.08ac | 65.80±0.23d |
| Enterobacter cloacae (OQ383316) | 61.53±0.23a | 71.77±0.12b | 49.93±0.15c | 54.53±0.54d |
| Bacillus cereus (OQ383317) | 63.00±0.38a | 80.77±0.12b | 58.33±0.38c | 51.60±0.31d |
| Alcaligenes faecalis (OQ383318) | 50.70±0.19a | 70.93±0.15b | 46.63±0.27c | 34.77±0.27d |
| Alcaligenes faecalis (OQ383319) | 64.53±0.15a | 75.10±0.27b | 61.23±0.50c | 39.50±0.19d |
| Bacillus subtilis (OQ383320) | 71.07±0.46a | 73.83±0.19b | 49.93±0.08c | 57.33±0.31d |
3.4. Effect of Different Carbon/Nitrogen Ratio
Generally, the tolerance indices of the isolates to test heavy metals at the respective carbon/nitrogen (C/N) ratios varied for the different metals, which could be a result of the different metabolic capabilities of the isolates; this observation was irrespective of heavy metals. Except for isolates C, E, G, and I, a significantly higher tolerance index to nickel was observed at C/N ratios of 5:2 and 5:3 for most of the isolates. However, significantly (p≤0.05), the highest tolerance index for lead was observed at a C/N ratio of 5:2 for isolates A, C, G, H, and J. For cadmium, significantly (p≤0.05), the highest tolerance index was observed at C/N ratios of either 5:5 or 5:4 for isolates A, C, E, F, H, I, and J. (Table 6).
In the case of nickel removal in the presence of the isolates, the significantly highest values were recorded in medium with a C/N ratio of 5:5, except for isolates B and J, where removal was observed to be significantly highest at CN ratios of 5:2 and 5:3, respectively. Similarly, the removal of lead in the presence of the isolates showed significantly higher values at C/N ratios of 5:5 or 5:4. For the removal of cadmium, the highest removal was observed at C/N ratios of 5:5 for isolates B, C, E, H, and J (Table 7).
| Bacterial Strains | C/N | |||
|---|---|---|---|---|
| 5:5 | 5:4 | 5:3 | 5:2 | |
| Nickel | ||||
| Yersinia enterocolitica (OQ383311) | 0.19±0.00a | 0.16±0.00b | 0.26±0.01c | 0.21±0.01d |
| Alcaligenes faecalis (OQ383312) | 0.14±0.00a | 0.19±0.00b | 0.28±0.00c | 0.45±0.01d |
| Bacillus cereus (OQ383313) | 0.21±0.01a | 0.32±0.00b | 0.20±0.00a | 0.17±0.03c |
| Alcaligenes faecalis (OQ383314) | 0.48±0.00a | 0.43±0.00b | 0.80±0.00c | 0.43±0.00b |
| Bacillus subtilis (OQ383315) | 0.29±0.00a | 0.20±0.00b | 0.21±0.00c | 0.23±0.00d |
| Enterobacter cloacae (OQ383316) | 0.34±0.00a | 0.26±0.00b | 0.35±0.01c | 0.31±0.00d |
| Bacillus cereus (OQ383317) | 0.46±0.01a | 0.19±0.00b | 0.20±0.00c | 0.21±0.00c |
| Alcaligenes faecalis (OQ383318) | 0.19±0.00a | 0.43±0.00b | 0.41±0.00c | 0.95±0.01d |
| Alcaligenes faecalis (OQ383319) | 0.68±0.00a | 0.49±0.00b | 0.49±0.00b | 0.57±0.01c |
| Bacillus subtilis (OQ383320) | 0.20±0.00a | 0.32±0.00b | 0.43±0.00c | 0.38±0.01d |
| Lead | ||||
| Yersinia enterocolitica (OQ383311) | 0.53±0.01a | 0.32±0.00b | 0.26±0.00c | 0.55±0.00d |
| Alcaligenes faecalis (OQ383312) | 0.75±0.01a | 0.66±0.01b | 0.60±0.01c | 0.62±0.01d |
| Bacillus cereus (OQ383313) | 1.16±0.02a | 1.35±0.00ab | 1.21±0.01a | 1.51±0.27b |
| Alcaligenes faecalis (OQ383314) | 0.56±0.00a | 0.77±0.00b | 0.55±0.00c | 0.61±0.00d |
| Bacillus subtilis (OQ383315) | 1.10±0.02a | 1.05±0.00b | 0.70±0.00c | 1.08±0.01a |
| Enterobacter cloacae (OQ383316) | 0.61±0.00a | 0.91±0.00b | 0.76±0.01c | 0.83±0.00d |
| Bacillus cereus (OQ383317) | 1.52±0.01a | 1.10±0.00b | 1.02±0.00c | 1.67±0.01d |
| Alcaligenes faecalis (OQ383318) | 0.96±0.01a | 0.78±0.00b | 0.68±0.00c | 1.01±0.01d |
| Alcaligenes faecalis (OQ383319) | 0.76±0.00a | 0.89±0.00b | 0.54±0.00c | 0.75±0.00d |
| Bacillus subtilis (OQ383320) | 0.54±0.00a | 0.79±0.00b | 0.58±0.00c | 0.95±0.00d |
| Cadmium | ||||
| Yersinia enterocolitica (OQ383311) | 0.12±0.00a | 0.07±0.01b | 0.07±0.00b | 0.09±0.00c |
| Alcaligenes faecalis (OQ383312) | 0.08±0.00a | 0.08±0.00ac | 0.06±0.00b | 0.08±0.00c |
| Bacillus cereus (OQ383313) | 0.32±0.01a | 0.16±0.00b | 0.17±0.00b | 0.22±0.04c |
| Alcaligenes faecalis (OQ383314) | 0.60±0.00a | 0.73±0.01b | 0.49±0.00c | 0.79±0.00d |
| Bacillus subtilis (OQ383315) | 0.23±0.01a | 0.32±0.01b | 0.30±0.00c | 0.18±0.00d |
| Enterobacter cloacae (OQ383316) | 0.50±0.00a | 0.72±0.00b | 0.56±0.00c | 0.21±0.01d |
| Bacillus cereus (OQ383317) | 0.67±0.00a | 0.58±0.00b | 0.60±0.00c | 0.72±0.00d |
| Alcaligenes faecalis (OQ383318) | 1.02±0.01a | 0.55±0.02b | 1.02±0.01a | 0.87±0.00c |
| Alcaligenes faecalis (OQ383319) | 0.70±0.00a | 0.48±0.00b | 0.41±0.00c | 0.36±0.01d |
| Bacillus subtilis (OQ383320) | 0.50±0.00a | 0.50±0.00a | 0.33±0.00b | 0.45±0.01c |
Table 7.
| Bacterial Strains | C/N | |||
|---|---|---|---|---|
| 5:5 | 5:4 | 5:3 | 5:2 | |
| Nickel | ||||
| Yersinia enterocolitica (OQ383311) | 74.10±0.27a | 68.63±0.27b | 59.10±0.12c | 67.20±0.15d |
| Alcaligenes faecalis (OQ383312) | 65.30±0.19a | 67.10±0.12b | 59.00±0.38c | 71.83±0.35d |
| Bacillus cereus (OQ383313) | 61.43±0.27a | 57.87±0.15b | 39.47±0.23c | 41.30±0.12d |
| Alcaligenes faecalis (OQ383314) | 74.63±0.27a | 50.33±0.38b | 64.47±0.23c | 48.33±0.23d |
| Bacillus subtilis (OQ383315) | 67.47±0.08a | 31.43±0.69b | 56.90±0.27c | 41.63±0.04d |
| Enterobacter cloacae (OQ383316) | 59.33±0.00a | 50.73±0.31b | 56.87±0.23c | 35.00±0.15d |
| Bacillus cereus (OQ383317) | 51.10±0.04a | 47.50±0.19b | 44.00±0.15c | 50.37±0.19d |
| Alcaligenes faecalis (OQ383318) | 59.30±0.19a | 46.27±0.31b | 45.60±0.23c | 50.33±0.38d |
| Alcaligenes faecalis (OQ383319) | 57.37±0.27a | 47.57±0.12b | 39.80±0.23c | 45.50±0.27d |
| Bacillus subtilis (OQ383320) | 47.50±0.12a | 43.60±0.08b | 49.23±0.12c | 36.40±0.15d |
| Lead | ||||
| Yersinia enterocolitica (OQ383311) | 83.03±0.27a | 77.10±0.27b | 72.17±0.19c | 68.33±0.23d |
| Alcaligenes faecalis (OQ383312) | 86.93±0.15a | 79.50±0.19b | 73.03±0.12c | 67.63±0.27d |
| Bacillus cereus (OQ383313) | 92.50±0.19a | 88.00±0.15b | 73.23±0.12c | 79.20±0.15d |
| Alcaligenes faecalis (OQ383314) | 84.90±0.12a | 64.83±0.19b | 73.77±0.27c | 59.37±0.27d |
| Bacillus subtilis (OQ383315) | 85.60±0.15a | 79.13±0.08b | 65.20±0.23c | 51.70±0.12d |
| Enterobacter cloacae (OQ383316) | 79.87±0.15a | 79.13±0.23b | 73.67±0.15c | 61.20±0.23d |
| Bacillus cereus (OQ383317) | 87.23±0.12a | 85.87±0.15b | 73.77±0.27c | 59.00±0.23d |
| Alcaligenes faecalis (OQ383318) | 79.43±0.27a | 82.33±0.38b | 73.03±0.12c | 60.93±0.31d |
| Alcaligenes faecalis (OQ383319) | 87.93±0.15a | 78.13±0.15b | 39.80±0.23c | 60.43±0.12d |
| Bacillus subtilis (OQ383320) | 79.10±0.27a | 80.37±0.12b | 77.03±0.04c | 67.30±0.19d |
| Cadmium | ||||
| Yersinia enterocolitica (OQ383311) | 77.77±0.12a | 83.53±0.23b | 78.90±0.19c | 88.33±0.38d |
| Alcaligenes faecalis (OQ383312) | 77.77±0.27a | 73.50±0.19b | 74.03±0.19c | 71.83±0.58d |
| Bacillus cereus (OQ383313) | 82.07±0.23a | 72.50±0.04b | 63.57±0.19c | 59.60±0.23d |
| Alcaligenes faecalis (OQ383314) | 74.97±0.12a | 73.93±0.08b | 63.97±0.27c | 75.43±0.27d |
| Bacillus subtilis (OQ383315) | 81.70±0.35a | 72.30±0.04b | 66.27±0.15c | 68.83±0.12d |
| Enterobacter cloacae (OQ383316) | 71.93±0.08a | 66.90±0.12b | 72.57±0.12c | 72.13±0.15d |
| Bacillus cereus (OQ383317) | 73.80±0.08a | 72.30±0.19b | 74.10±0.12a | 77.00±0.38c |
| Alcaligenes faecalis (OQ383318) | 69.87±0.15a | 66.57±0.12b | 65.93±0.08 | 60.60±0.23d |
| Alcaligenes faecalis (OQ383319) | 65.00±0.38a | 70.97±0.19b | 62.33±0.08c | 59.20±0.15d |
| Bacillus subtilis (OQ383320) | 70.83±0.04a | 67.07±0.08b | 63.07±0.46c | 61.00±0.23d |
4. DISCUSSION
The bacterial strains in this study showed significantly high tolerance to the test metals in media containing sodium acetate as a carbon source. With respect to the removal of heavy metals, higher removal efficiencies were reported in media that contained sucrose as a carbon source in the presence of most of the isolates. The higher removal efficiency of the metals observed in the presence of sucrose as a carbon source could result from its complexation capacity. It is hypothesized that since sucrose is a complex and larger molecule, it possesses more functional groups that enable complexation, which enables adequate binding to the metal ions that may ease metal removal by a microbe [10]. A similar observation has been reported by earlier workers [11]. The addition of external carbon sources is reported to have improved the biological nutrient removal processes, as the type of carbon added showed a different removal pattern [11]. In addition, previous investigators have reported sucrose as a carbon source that is easily utilized by microorganisms [12].
Similarly, the use of acetate as a preferred carbon source has been reported in related studies [11, 13, 14]. Another previous study [15] reported that high tolerance and high removal efficiency by Acinetobacter sp. was observed in media grown with sodium pyruvate, sodium citrate and sodium acetate when used as a carbon source; the study also reported low removal efficiency in media containing sucrose and glucose. The different variations in tolerance and removal patterns of the isolates in the respective carbon sources may be due to their metabolic and biochemical capabilities [16]. It is indicated that the absence of a carbon source resulted in no positive influence on the removal or reduction of chromium [16].
In a study on the effects of carbon sources on Cr (VI) reduction [17] in the presence of P. aeruginosa AB93066 in nutrient broth, the order of preference was glucose > glycerine > butyric alcohol > citric acid > sodium acetate >oxalic acid > lactose > sucrose > methanol > and phenol. It is hypothesized that since glucose is a readily oxidized carbon source, it could serve as a good electron donor [18]. Moreover, a related study on heavy metal tolerance and removal by Acinetobacter sp. SCYY-5 [19] reported that the preferred order of tolerance to metals at different carbon sources was citrate, followed by soluble starch > glucose > fructose > lactose > sucrose.
In this study, potassium nitrate was observed as an ideal nitrogen source for tolerance to the test metals, while the highest removal efficiencies were observed in media that contained yeast extract in the presence of the majority of the isolates. The higher tolerance but lower removal efficiency of the metal observed in media with potassium nitrate could be attributed to the prioritization of the production of energy from nitrate reduction over metal removal. It could also be attributed to decreased binding of nitrate ions to the metal, which could lead to decreased metal removal [20]. Meanwhile, high removal efficiency in media containing yeast extract could probably be a result of yeast extract being readily utilized as an organic nitrogen source by the isolates, unlike potassium nitrate, which requires the nitrate reductase enzyme in converting nitrate into ammonium for the metabolism of nitrogen [21]. Furthermore, yeast extract has been found to contribute to the production of extracellular polymeric substances (EPS) by bacteria, thereby enhancing heavy metal removal efficiency due to the EPS binding to the heavy metal ions [22]. Previous studies have also shown a 25%-50% reduction in Cr (VI) when grown in yeast extract media [16].
In a study by Murugavelh and Mohanty [19], a 96.7% reduction of Cr (VI) was reported when yeast extract was used as a nitrogen source at a concentration of 5 g/L in media. However, another study reported ammonium nitrate as the optimal nitrogen source compared to other nitrogen sources, such as potassium nitrate, yeast extract, peptone, urea, and sodium nitrate [23]. Similarly, a related study [24] reported the highest lead and cadmium removal by Stenotrophomonas koreensis in media that contained potassium nitrate as a nitrogen source. Other authors have reported the nitrogen utilization rate to be ammonium chloride < ammonium sulfate < potassium nitrate < yeast extract < L-glutamic acid [18].
With respect to the C/N ratio, variations were observed in the tolerance indices of the isolates to the test metals. Higher metal removal was observed in media with C/N 5:5. In a study [25] on the heavy metal remediation potential of landfill soil bacterial isolates, it was indicated that medium supplemented with carbon source concentrations of 4-6 g/L enhanced heavy metal remediation potential of Klebsiella edwardsii, Pseudomonas aeruginosa and Enterobacter cloacae. It is reported [26] that a lower C/N ratio favors removal by isolates as they consume carbon more than nitrogen. Xu et al. [28] indicated that higher metal removal efficiencies were recorded
at a C/N ratio of 5:1. It is opined [27] that a higher C/N ratio could stimulate intense biological metabolism. Existing studies also state that the C/N ratio varies as it depends on the nature of the species and their ability to metabolize the nitrogen source in a medium [21].
In a related study by Ali et al. [29], a C/N ratio of 3:1 was reported as optimum for metal removal. The carbon/nitrogen ratio is reported to influence the composition and concentration of metabolites in bacteria and also influence the metabolic capabilities of microbes [30-32]. Yuncu et al. [33] indicated that the heavy metal sorption capacity varied with the C/N ratio, which could result from the different biosorption capabilities of the microorganisms used. A lower C/N ratio increased the biosorption capacity of Cu (II), while a high C/N ratio increased the biosorption capacity of Cd (II). The study further reports that the lowest and highest Zn (II) capacities were observed at a C/N ratio of 4:3 and 2:1, respectively [33].
CONCLUSION
From the findings of this study, 10 out of the 65 bacterial strains isolated showed significant tolerance and removal to the test metals lead, cadmium and nickel. The extent of heavy metal tolerance and removal potential in the presence of the test isolates depends on the carbon source, nitrogen source, and C/N ratio. Although tolerance to metals was observed when the different carbon sources were used, the significantly highest tolerance index was recorded in media that contained sodium acetate as the carbon source in the presence of most of the isolates. The significantly high tolerance to sodium acetate was probably due to the production of metal-chelating substances.
In the case of nitrogen sources, the highest tolerance to the test metals was observed in media containing potassium nitrate in the presence of most of the isolates. However, the significantly highest lead removal was observed in media containing potassium nitrate, while cadmium and nickel removal was significantly highest in media containing yeast extract. Generally, C/N ratios did not follow any visible trend with respect to tolerance and removal of metals in the presence of the bacterial species. This, therefore, shows that the effects of carbon and nitrogen sources, the C/N ratio in remediation of heavy metal, depend on the isolate type and the heavy metal. The effects of these factors directly influence the metabolic pathways, enzymatic activities and growth rate of the bacterial test species, thus enhancing the biosorption capacities of the test metals. Overall, the potential of the isolates in the remediation of heavy metal-polluted environments could be further exploited and enhanced by carefully selecting these factors, thereby contributing to environmental sustainability.
LIST OF ABBREVIATIONS
| AAS | = Atomic Absorption Spectrometry |
| TI | = Tolerance index |
| NCBI | = National Centre for Biotechnology Information |
| ANOVA | = Analysis of Variance |
| EPS | = Extracellular polymeric substances |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
All the data and supportive information is available within the article.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors are grateful to Afe Babalola University for providing facilities for the study.


