All published articles of this journal are available on ScienceDirect.
Current Status and Future of Nitrile Catalysis using Key Nitrilases Enzymes and their Biotechnological Impact
Abstract
Nitriles are organic compounds consisting of −C≡N group. They are frequently known to occur in nature and as intermediate by-products and waste products of various chemical, pharmaceutical, and agricultural industries. They are also found in fruit pits, cabbage, cauliflower, and sprouts, which are released upon hydrolysis. Nitrile converting enzymes like nitrilases have been extracted from microorganisms and plants. Nitrilase-mediated biocatalysis reactions have continuously aroused widespread interest to scientists and entrepreneurs in organic synthesis. Nitrile converting biocatalysts (Nitrilases) are now of substantial industrial interest from the perspective of treating toxic nitrile and cyanide-containing compounds. Nitrile degrading enzymes generally consist of nitrilases and amidases. The aim of the current review is to summarize the recent advancements on regioselective nitrilases concerning their fundamental researches and their application in the synthesis of series of high-value fine chemicals and pharmaceuticals. The present review also focuses on the utility of nitrile converting enzyme, sources, properties, classification, structure, and applications as well.
1. INTRODUCTION
Chemical reactions occur either spontaneously or require a catalyst to move in a forward direction. Catalysts are molecules that reduce the magnitude of the energy barrier required to achieve a transition state for a substance to be transformed into a product [1]. Catalysts can be organic or inorganic in nature. Enzymes are the catalysts which bind to the substrate and transform it into subsequent product thus called Biocatalysts and the process as Biocatalysis. Emil Fischer proposed (1894) the “lock and key” model for explaining the mechanism of enzyme catalysis, and Buchner (1897) demonstrated that cell-free extracts were capable of catalyzing fermentation processes. In the 20th century, the research and development on enzyme catalysis grew rapidly along with their industrial and biotechnological application. Enzymes are remarkable catalysts capable of catalyzing various chemical reactions in the cell. Enzymes are selective for their substrates and are also known to determine different metabolic pathways. Furthermore, with the help of biotechnological tools, better strategies and variants can be engineered, which can ease off the economic burden as well as provide better product yields.
Enzymes are also referred to as green catalysts and have an ability to transform a broad range of complex molecules with additional advantage of entire process being more chiral (enantio-) and positional (regio-) selective. Furthermore, it does not require any complicated blocking and deblocking steps, which are essential in chemical catalysis. Enzymes have also been exploited for the transformation of complex organic compounds as well as some of the inorganic compounds which are toxic or difficult to transform by chemical catalysis [2, 3]. Nitrilases (EC 3.5.5.1), an enzyme of the nitrilase superfamily, could catalyze nitrile-containing compounds into their corresponding carboxylic acids and ammonia in a single step reaction [4]. The earliest applications of nitrilase enzymes were a synthesis of optically pure amino acids by utilizing acylases, hydantoinases, and formation of bulk chemical acrylamide from acrylonitrile using nitrile hydratases [4]. Nitrile compounds are generated as intermediate products by-products, and waste products of agriculture, chemical, pharmaceutical, and processing in fossil and fuel industries [5, 6]. In general, nitriles are simple to synthesize using various chemical pathways such as the addition of cyanide to alkyl halide. However, nitriles are known to be toxic, mutagenic, and carcinogenic because of their cyano (-C≡N) group. Furthermore, the regular methods of removing waste by combustion and dumping have proven to be ineffective, and in fact, has led to the production of even more toxic cyanide compounds. In order to avoid this problem, bioremediation strategies have been employed for the treatment of such chemicals. Many microbes are able to hydrolyze nitrile to the less toxic and valuable carboxylic acid by using nitrile-utilizing enzymes. These enzymes are known to have an important role in nitrile degradation. In recent years nitrilase-mediated catalysis has attracted substantial attraction. Various nitrilase-producing organisms, including bacteria, fungi, yeast, and plants, have been described. Such microbial cell factories can be utilized for the commercial production of carboxylic acid. The success of the commercial production of nicotinic acid and mandelic acid using nitrilase-mediated catalysis has shown the great economic potential of this enzyme [7].
The present review focuses primarily on advancements in research with reference to the nitrilase production, characterization, biochemical and biotechnological applications of the enzyme. We have also discussed the origin and environmental distribution of nitrile compounds with different bioremediation pathways and enzymes involved therein.
2. NITRILASE SUPERFAMILY
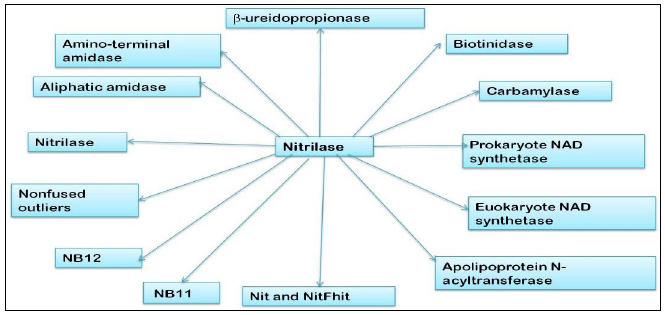
Wide varieties of microorganisms, plants, and animals are capable of performing nonpeptide carbon-nitrogen hydrolysis reactions using enzymes belonging to the nitrilase superfamily. Many bacteria and archaea, particularly those with an ecological relationship to plants and animals, encode members of the nitrilase superfamily. They utilize the enzymes for chemically similar nitrile or amide hydrolysis reactions or the condensation of acyl chains to polypeptide amino termini. On the basis of sequence similarity and the presence of additional domains, the superfamily can be classified into 13 branches, nine of which have known or deduced specificity for specific nitrile- or amide-hydrolysis or amide-condensation reactions [8]. The 13 branches of nitrilase superfamily are (Fig. 1) i) nitrilase, ii) aliphatic amidase, iii) amino-terminal amidase, iv) biotinidase, v) b-ureidopropionase, vi) carbamylase, vii) prokaryote NAD synthetase, viii) eukaryote NAD synthetases, ix) apolipoprotein N-acyltransferase, x) Nit and NitFhit, xi) NB11, xii) NB12, and xiii) nonfused outliers. Within the nitrilase superfamily, nitrile hydrolyzing enzymes are from two distinct groups, nitrilases (EC 3.5.5.1) which belong to 3rd class “hydrolases,” and nitrile hydratases (EC 4.2.1.84) belonging to the 4th class “lyases”. A group of amidases (EC 3.5.1.4) that actively participates in the hydrolysis of amides are also related to the nitrilase/cyanide hydratases family [9].
3. NITRILE METABOLISM
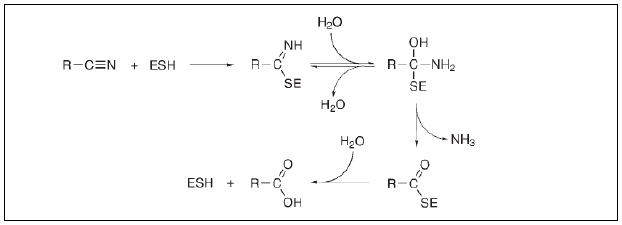
Nitrile compounds are considered highly toxic, and upon metabolism, it produces even more toxic cyanide compounds. First-ever account of nitrile metabolism by nitrilase was reported by Thimann and Mahadevan [10] in plants. Later similar activity was observed in bacteria by Robinson and Hook [11]. Harris et al. [12] first gave a detailed account of how nitrile and cyanide compounds are introduced in the environment and metabolized by living organisms. The main sources for cyanide and nitrile compounds are Industrial wastes/ by-products and biological activity (mostly in plants). Furthermore, it has been reported by Harris et al. [12] that nitriles and cyanide compounds are degraded by β-Cyanoalanine synthase, Cyanide hydratase, Rhodanase, Nitrogenase, cyanide oxygenase enzymes. Nagasawa et al. [13] discovered novel nitrilase enzyme-producing bacteria Alcaligenes faecalis, which showed its commercial viability. All these enzymes were later included in the nitrilase superfamily. The core mechanism of nitrile compounds metabolism in microorganisms was by means of nitrilase and nitrile hydratase activity. The mechanism involves a nucleophilic attack on the nitrile C-atom by a conserved cysteine residue of the nitrilase, producing NH3 with the first water molecule addition, and yielding acid and a regenerated enzyme with the addition of the second water molecule (Fig. 2) [14].
4. SOURCES OF NITRILASE
Since the discovery of nitrilase in the 21st century [10], many researchers have been fascinated by its unique properties. Initially reported from plants, now the nitrilase enzyme has been discovered from many fungi, bacteria, and yeast [15]. Various potential nitrilase producer organisms are listed in Table 1.
| Bacteria | Plants | Fungi |
|---|---|---|
|
Acido vorax facilis 72W Acinetobacter sp. (strain AK226) Alcaligenes faecalis ATCC 8750 Alcaligenes faecalis JM3 Bacillus sp. strain OxB-1 Bacillus pallidus Dac521 Comamonas testosteroni Klebsiella pneumoniae ssp. Ozaenae Pseudomonas sp. (SI) Pseudomonas DSM 7155 Rhodococcus NCIB11215 Rhodococcus K22 Rhodococcus ATCC39484 Rhodococcus rhodochrous JI |
Arabidopsis thaliana Barley Chinese cabbage |
Fusarium oxysporum Fusarium solani IMI196840 Penicillium multicolor Aspergillus niger K10 |
Bacteria have been the prime focus for nitrilase production by researchers due to their growth rate and high yields of enzyme production. The first ricinine nitrilase was discovered in the Pseudomonas genus [17]. Later nitrilase enzyme was discovered in many other bacterial species such as Rhodococcus, Nocardia, Acinetobacter, Alcaligenes, Bacillus, Corynebacterium, etc. [18-20]. Recently researchers have focused on the isolation of novel bacterial strains from the extremophilic environment in order to find unique nitrilase which could have the better catalytic potential [21].
Fungi and yeasts have also been exploited for remediation purposes from time to time and are also known to survive in harsh environment [22]. The first fungal nitrilase was reported from Fusarium solani [23]. Several scientists have reported properties and production of nitrilase from various other fungal genera viz. Fusarium, Gibberella, Aspergillus, and Penicillium [22, 24-26]. Many yeast species have been reported for their nitrile compound degradation capabilities, but most of them are by means of nitrile hydratase and other enzymes, and only a few species have been identified, which are nitrilase enzyme producers [25]. Some of the noteworthy nitrilase producer yeasts are Torulopsis candida [27], Exophiala oligosperma R1 [28], and Pichia pastoris [29].
5. FACTORS AFFECTING NITRILASE
In order to achieve maximum productivity, the enzyme must be provided with an optimal environmental condition and efficient feeding of the substrate. Nitrilases are well known for their high specificity and sensitivity toward different substrates. Factors affecting nitrilase activity are discussed below
5.1. Nutrients in Media
Enzyme productions, as well as activity of enzymes, are highly influenced by media components. Among the contents of media, carbon and nitrogen sources play a significant role in enhancing cell proliferation and nitrilase biosynthesis [30]. Furthermore, in some cases, it was observed that the type and amount of these sources have a regulatory response toward the synthesis of nitrilase by a mechanism such as catabolic repression. For example, in P. putida, MTCC 5110 glycerol acted as a catabolic repressor. Although it supported the higher biomass production, enzyme yields were lower [31]. So far, various studies have confirmed that Sodium acetate, starch, glucose, and mannitol are better carbon sources, whereas peptone, yeast extract, and casamino acid are better nitrogen sources for nitrilase production in Alcaligenes sp., A.faecalis, A. nitroguajacolicus, P. putida, Rhodococcus sp., and E. oligosperma [32-34].
5.2. Presence of Inducers, Repressors, and Modifiers
Most of the nitrilases are reported to be inducible by substrates, products, and their analogs, but not much information is available regarding constitutive nitrilases, besides those of Brevibacterium R312, K. ozaenae, B. subtilis ZJB-063, and Alcaligenes sp. ECU0401 [33, 35, 36]. In general, certain nitriles, amides, and carboxylic acids are known to be nitrilase inducers. For example, ε-Caprolactam was found to be a powerful inducer of the R. rhodochrous J1 nitrilase. It is also responsible for an effective induction in some other Rhodococcus strains [37]. Rhodococcus sp. NDB 1165 nitrilase was found to be inducible by propionitrile [38]. Benzonitrile has been reported as an inducer for nitrilase from P. aeruginosa 10145, in which cell growth was inhibited by the addition of benzonitrile at the beginning, and thus the addition of an inducer was made after 24 h cultivation [39]. The effects of different inducers on the substrate specificity of A. faecalis MTCC 10757 has been studied, and acrylonitrile was proven to be the most preferred inducer and substrate [32].
Although not extensively studied, several enzyme modifiers for nitrilases have been reported. The catalytic site of nitrilase possesses cysteine residues, and thus the thiol binding agent affects its activity negatively. Nitrilase activity is strongly inhibited by thiol-binding reagents, such as N-ethylmaleimide, 5, 5-dithiobis (2-nitrobenzoic acid), iodoacetamide, iodoacetic acid, p-chloromercuribenzoate, and p-hydroxymercuribenzoate [31, 35, 40, 41]. Nitrilases do not require metal ion as a cofactor, so it is mostly proven to be unaffected by metal chelating agents like Ethylene Diamine Tetraacetic Acid (EDTA), sodium azide, 8-hydroxyquinoline, and o-phenanthroline [42, 43].
5.3. Effect of Physiological Condition
Enzymes being proteinaceous in nature are sensitive to both temperature and pH changes. Temperature and pH often affect the structure and the nature of an enzyme, thereby further influencing the enzyme activity. Similar to other enzymes nitrilase also work optimally under the mild condition with pH ranging 7 to 8. However, certain nitrilases have been reported to be optimally active at moderately acidic and alkaline pH. Jandhyala [14] reported nitrilase from Bacillus pumilus C1 and Pseudomonas stutzer showing high activity even at pH 5.0 - 6.0. Nitrilase activity has been reported in the temperature range 30°C to 55°C. However, thermoactive nitrilase has also been reported from the anaerobic archaea P. abyssi which grows optimally at 100°C [40]. Dennett et al. [21] reported thermostable nitrilase from Pyrococcus sp. M24D13 showed optimal activity at 85°C and pH 7.5.
The enzyme works efficiently under optimal conditions; however, its activity is inconsistent because it not always in stable condition. Stability is one of the characteristics which determines the commercial viability of an enzyme. The nitrilases from P. putida, Microbacterium paraoxydans, M. liquefaciens [44], A. nitroguajacolicus [43], and A. facilis [45] had half-lives of 27.3 h, 1.7 h, 5.4 h, 157.6 h, and 14 d at 30°C, respectively. Remarkably, a highly thermostable nitrilase, with a half-life of 25 h at 70°C, 9 h at 80°C, and 6 h at 90°C was reported from P. abyssi [40]. Bhatia et al. [46] were able to retain 80% activity at 4 °C for 10 days and 50% activity after 7 reuse cycles using chitosan microsphere.
5.4. Effect of Organic Solvent
Organic solvents have been recognized to increase the solubility of substrates or products in the bioconversion reaction. The enzymatic reactions in an organic solvent, such as the lipase-mediated reactions, are well known. Similarly, nitrilases have interesting characteristics of being tolerant to several organic solvents; for example, Heinemann et al. [47] observed that the presence of methanol increased the rate of reaction of the recombinant nitrilase from Synechocystis sp. PCC6803. The maximal conversion of this nitrilase was achieved in the presence of 40% (v/v) hexadecane. More than half of its initial activity was maintained in the presence of 5-50% of n-hexane and n-heptane, 5-15% of xylene and ethanol. Layh et al. [48] reported activity of Pseudomonas sp. DSM 11387 to be retained (60-109%) in the presence of 50% (v/v) hexadecane. In addition, the presence of large amounts of organic solvents in the oxynitrilase mediated transformation reaction was reported to suppress the reversible chemical reaction [49]. The organic solvent increased the tendency of the chemical equilibrium to synthesize more products.
5.5. Structural Configuration and Genetic Engineering
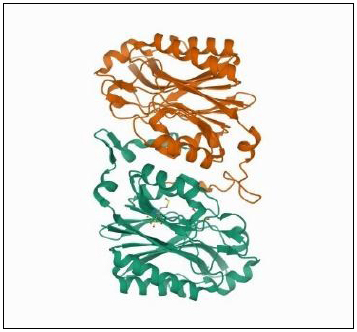
Enzyme activity is greatly affected by the 3D configuration of the enzyme. Any changes in helical structure can impact either positively or negatively. Woodward et al. [50] reported that plant nitrilases are oligomeric helix forming enzymes in nature, and any change in their helical twist can alter the specificity of the enzyme as binding pocket residue is found inside the helical twist. Investigations revealed that altering the α2 and α3 loops of the binding pocket in the helix led to enzymes accepting the substrates in which nitrilases are conventionally do not interact [50]. Fig. (3) represents the crystal structure of hyperthermophilic nitrilase from Pyrococcus abyssi GE5 expressed in E. coli.
It is a widely known phenomenon that introducing mutations in enzymes and heterologous expression can improve the production as well as activity of enzymes. Several investigations have proven that the efficiency and kinetics of nitrilase can be improved by utilizing these technologies [31, 51-53].
6. CLONING, EXPRESSION, AND ENGINEERING OF NITRILASE
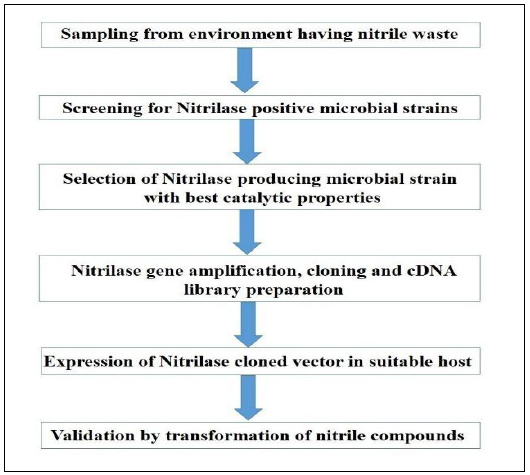
Although highly efficient enzyme-producing microbes are found in nature, the catabolic properties and kinetic parameters can be further improved by the assistance of molecular biology tools. Constructing a recombinant enzyme provides an opportunity to meet the application of natural biocatalysts with high efficiency. Fig. (4) depicts the general strategy for the heterologous expression of nitrilase.
Nitrilase gene has been heterologously expressed, and the first nitrilase gene cloned in Escherichia coli encoded a bromoxynil-degrading activity from Klebsiella pneumonia subsp. Ozaenae [36]. Bartling et al. [54] expressed the first plant gene from Arabidopsis thaliana. The nitrilase gene cloned from Alcaligenes faecalis JM3 by Kobayashi et al. [55] showed a 35% increase in the specific activity for thiophene-2- acetonitrile. Chauhan et al. [56] cloned regioselective aliphatic nitrilase from A. facilis 72W and overexpressed under the control of strong T7 promoter in the pET-3c vector. The engineered nitrilase displayed a 15-fold higher enzyme activity than the native strain. The thermostable recombinant nitrilase isolated by Mueller et al. [40] from Pyrococcus abyssi has proven highly thermostable, with a half-life of 6 h at 90°C. Other recombinant nitrilases are from Rhodococcus rhodochrous PA-34 [51], Rhodococcus rhodochrous tg1-A6 [57], Alcaligenes faecalis ZJUTB10 [58], Synechocystis sp. Strain PCC6803 [47], Alcaligenes sp. ECU0401 [52], Gordona terrae MTCC-8139 [59] have been cloned. Aromatic nitrilase was cloned from Pseudomonas putida CGMCC3830 by Zhu et al. [60]] and it showed high specificity towards aromatic nitriles such as 3-cyanopyridine, 4-cyanopyridine, and 2-chloro-4-cyanopyridine. Wang et al. [61] cloned a novel nitrilase gene from Rhodobacter sphaeroides LHS-305 and overexpressed it in Escherichia coli. These nitrilases have a very good prospective for commercializing the synthesis of various cyanocarboxylic acids by hydrolyzing readily available dinitriles. A high (R)-enantioselective nitrilase gene from Sphingomonas wittichii RW1 cloned and overexpressed in E. coli showed 95% enantiomeric excess, which made it a promising biocatalyst for the synthesis of (R)-phenylglycine by Qiu et al. [62]. Among fungal genera, heterologous expression, purification, and characterization of nitrilase gene from Aspergillus niger K10 [63] and Fusarium proliferatum [64] has been reported. Later Kaplan et al. [63] utilized a genome mining tool to explore more such fungal nitrilases and synthetic genes of putative nitrilase from Gibberella, Neurospora, and Aspergillus were expressed in E. coli by Gong et al. [65]. Most of them are based on cloning nitrilase genes under the T7 promoter (controlled by the lac operator), mainly in the pET plasmids, and their induced expression in Escherichia coli (DE3) strains to transcribe the T7 RNA polymerase gene from the lacUV promoter inducible by Isopropyl-b-Dthiogalactopyranoside (IPTG). This system was used for the over-expression of many bacterial [52, 58, 66] as well as fungal [64, 65, 67] nitrilase genes. Seth et al. [68] were able to isolate Rhodococcus pyridinivorans NIT-36, an alkalophilic nitrilase-producing strain capable of transforming both aliphatic and aromatic nitriles from hot-water springs of Tattapani, Himachal Pradesh, India, and found the highest activity and thermostability for agar immobilized cells, and they retained 70% activity even after 5 h. Singh et al. [69] were able to isolate a broad spectrum nitrilase from moderately halophilic bacterium Halomonas sp. IIIMB2797. Agarwal et al. [70] were able to achieve nitrilase production of 11.54 U/mg using benzonitrile as an inducer and were able to further improve it by applying optimization study of response surface methodology. Thakur et al. [53] produced an aryl-specific inducible nitrilase from Alcaligenes faecalis MTCC 12629 which hydrolyzed 4-aminophenylacetonitrile to 4-amino phenylacetic acid. Bacillus subtilis strains have been demonstrated to be suitable heterologous producers of nitrilases, especially for practical purposes owing to their spore-forming and biofilm-forming abilities. B. subtilis spores were used as a matrix for the immobilization of nitrilases from the thermophilic bacteria Clostridium thermocellum [71] and Thermotoga [72]. In one of the most noteworthy research multicopy integrations of nitrilase gene in Pichia under the control of the Alcohol Oxidase 1 (AOX1) promoter and co-expression of ER Oxidoreductin 1 (ERO1) resulted in an 18-fold increase in the nitrilase activity [29]. Furthermore, its kinetic parameters were also improved.
7. APPLICATIONS OF NITRILASE
In recent times, the interest in biocatalysis and biotransformation has been constantly growing because of their environmentally benign properties and high selectivity. Initially, nitrilases were used specifically for the production of carboxylic acids from naturally occurring nitrile compounds such as mandellonitrile and indoleacetonitrile. In recent years, nitrilases have been used for the production of novel organic compounds with medicinal properties, chemical intermediates production, and bioremediation also. Major applications of nitrilases are discussed below.
7.1. Biotechnological Potential of Nitrilase
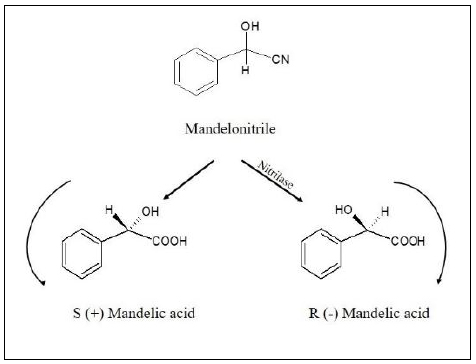
Green chemistry is a bridge between chemistry and biology where the main goal is to replace chemical reactions with biocatalysis in order to reduce or even eliminate harsh chemicals used in the process [25]. Nitrilase enzyme is one good example of green chemistry being put to practical purpose. The biotechnological applications of nitrilase range from detoxification of cyanide and nitrile present in the environment to the production of bioactive compounds [73]. Plants produce a variety of nitrile and cyanide compounds such as phenylpropionitrile and β-cyano-L-alanine as a part of their defense mechanisms, where nitrilases can be employed to transform and produce plant-specific compounds [74]. Nitrilases are best known for the production of plant hormone indole -3 acetic acid [55]. Apart from it, plants contain many glycosonitrile and liponitrile compounds, which are yet to be investigated for their bioactive properties [75]. Nitrilases have also been employed in the production of pharmaceutically important carboxylic acids from nitriles such as mandelonitrile, glyconitriles, etc. [25]. One of the important properties of nitrilase is its stereospecificity. For example, nitrilases are excellent producers of Stereospecific (R)-(−)-mandelic acid (Fig. 5). Overall, Nitrilases have great biotechnological potential, and many are naturally occurring nitrile compounds that can be of interest for the generation of biologically interacting molecules by means of biocatalysis.
7.2. Organic Acid Production
Nitrilases have been successfully commercialized for the production of organic acids by converting nitrile compounds. Many such nitrile compounds are either naturally occurring or synthesized organically. Nitrile conversion to organic acid requires the use of toxic chemicals, while nitrilase does it in a single step by simply hydration reactions. It is a total environmentally benign approach. Some of the important products are Nicotinic acid, Glycolic acid, Indoleacetic acid, Mandelic acid, and many more.
Nicotinic acids are also known as Vitamin B3. As shown in Fig. (6), nitrilases can efficiently replace the conventional method of preparing nicotinic acid from the oxidation of extracted nicotine. Nicotinic acids have a wide range of pharmaceutical applications [76]. First-ever nitrilase-assisted conversion of nicotinonitrile was achieved in Rhodococcus rhodochrous J1, which catalyzed direct hydrolysis of 3-cyanopyridine to nicotinic acid [5]. The most recent development in enzymatic production of nitcotinic acid improvement of the process by utilizing Sodium Alginate-Polyvinyl Alcohol (SA-PVA) immobilized cells of Pseudomonas putida mut-D3 expressing nitrilase [77].
Mandelic acid is another important organic acid which has a broad range of applications and one of the most common way to synthesize it, is by means of nitrilase enzyme (Fig. 6). This method has been commercialized and is notable due to its enantioselectivities of the reaction. The most popular method till today is overexpression of the Nitrilase from Alcaligenes sp. in E. coli, which resulted in the specific activity of 7000 U·L-1 toward (R,S)-mandelonitrile, and approximately 520 mM (R)-(−)-mandelic acid was accumulated after 17.5 h of conversion with 600 mM (R,S)-mandelonitrile in a fed-batch reaction [78].
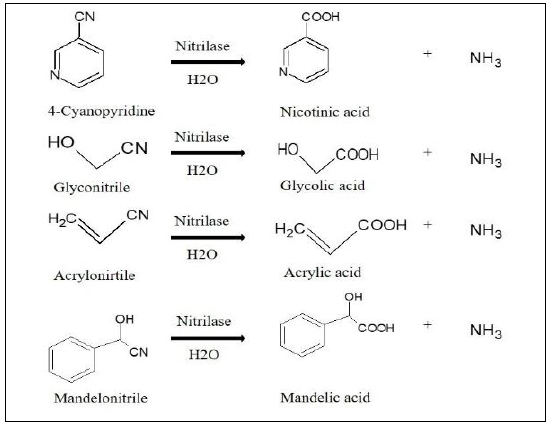
Apart from nicotinic acid and mandelic acid, acrylic acid and glycolic acids are also popular products that are produced by utilizing nitrilase enzyme (Fig. 6). Acrylic acids are also known as the simplest unsaturated acid, and their main application lies in the generation of acrylic salts and polyacrylic compounds. One of the first reported methods is the production of acrylic acid and methacrylic acid from acrylonitrile and methacrylonitrile by ε-Caprolactam-induced Rhodococcus rhodochrous J1. A total of 390 g acrylic acid/l and 260 g methacrylic acid were reported [79]. Recently biotransformation of acrylamide to acrylic acid was achieved from novel microorganism Bacillus tequilensis (BITNR004), and biotransformation efficiency of 63% and 59% obtained at 1 mM and 2 mM acrylamide, respectively. Glycolic acids belong to the α-hydroxycarboxylic acids class, and their major application lies as household cleaning, Cosmetics, industrial and electronic elements, vehicle oil additives, Liquid flow enhancers, pH controlling, and many others [80]. In one of the earlier reports, 96.5% yield of glycolic acid was obtained after 14 h of biocatalysis from a total of 200 mM glycolonitrile from Alcaligenes sp. ECU0401 in a lab scale fermenter [33].
7.3. Commercial Applications of Nitrilase
Precursors of many of the industrially important compounds are found in nature, but it requires a lot of processing to obtain active components of it. Furthermore, it is important to get the desired compound in its stereospecific forms, which lead to more complications. Nitrilases specifically target the nitrile group regardless of the organic moiety attached to it and transform it enantioselectively. Some of the important such intermediate compounds are β- Amino-amides, Cyanocyclohexylacetic acid, etc.
β- Amino-amides and acids can be used as building blocks for the synthesis of pharmaceutical intermediates, and nitrilases are capable of synthesizing such compounds from their nitrile precursors. This includes biologically active peptides and small molecule pharmaceuticals [81]. β -Amino acids are constituents of compounds such as the antitumour drug Taxol [82], the antifungal antibiotic Cispentacin [4], and the antidiabetic drug Sitagliptin [83]. Another pharmaceutical application is for inclusion in peptidomimetics that may be of use as protease inhibitors against retroviruses such as HIV [84]. Recently “super nitrilase mutant” of nitrilase with high activity, thermostability, and improved product tolerance from Acidovorax facilis ZJB09122 was expressed in E. coli for the conversion of a high concentration of 1-cyano cyclohexylacetonitrile-to-1-cyanocyclohexaneacetic acid, which is an important precursor of gabapentin. It resulted in improving all the kinetic parameters of the nitrilase enzyme. High substrate concentrations such as 1.3 M, 1.5 M, and 1.8 M were hydrolyzed by 13.58 g DCW/L with the productivity of 1-CA around 740 g/L/day [85].
7.4. Bioremediation Potential of Nitrilases
Cyanide and nitrile compounds occur naturally and are also at an industrial scale. These compounds are produced as a by-product or a waste of certain chemical industries. Nitrile wastes are particularly of concern because of their extreme toxicity. Nitrile-containing wastes are difficult to be degraded in nature, and physico-chemical methods are environmentally hazardous. In fact, incomplete chemical treatment or physical treatment such as incineration often leads to bigger problems such as the production of toxic hydrogen cyanide fumes, accumulation of cyanides, etc. Thus, it has become essential to remove nitriles and cyanides from the environment by means of environmentally friendly processes. Nitrilase class of enzyme specifically acts on cyanide and nitriles groups and has the unique characteristic of tolerating organic solvents. Therefore, Bioremediation of cyanide and nitrile waste by means of nitrilase and nitrilase producing organism can be an effective strategy for it. Fig. (7) represents various modalities for the remediation of nitrile and cyanogenic wastes.
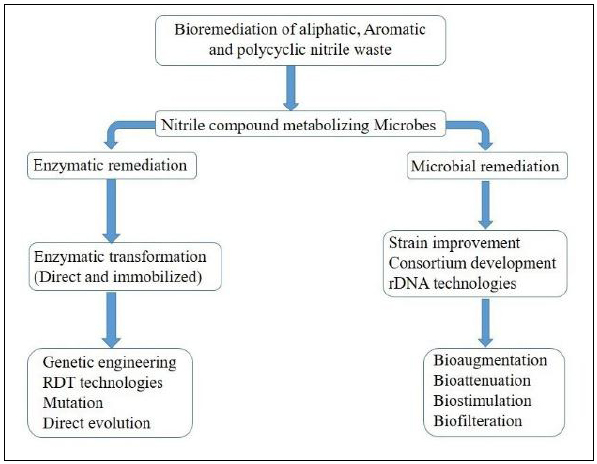
It is known that plenty of microorganisms are able to transform simple forms of cyanide. One of the earliest attempts of Bioremediation of cyanide waste was by means of Pseudomonas fluorescens NCIMB 11764; they were able to detoxify potassium cyanide waste [86]. Later a bacterial strain Paracoccus sp. SKG was isolated, which was capable of utilizing acetonitrile as a sole source of carbon, and nitrogen was isolated from the chemical waste samples [87]. In one of the important discovery, metallotolerant Pseudomonas pseudoalcaligenes CECT5344, isolated from sludge, was effective in utilizing waste from metal and jewelry industry waste [88]
Cyanide compounds are known to form a complex with metal ions, thus have been rigorously used in mines for metal extractions which creates a problem of accumulation of cyanides in the environment. Bioremediation of cyanide might be an efficient, cost-effective, eco-friendly, and attractive alternative to conventional physical and chemical waste processing [89]. Recently an efficient nitrile-degrading bacterium, Rhodococcus rhodochrous BX2, was isolated, which is being explored for the remediation capabilities of contaminated sites containing nitrile waste from industries [90].
Nitrile and cyanide wastes have been often ignored by researchers due to more emphasis on other pollutants. Although found in a lesser amount – C≡N containing wastes are extremely hazardous and have a long-term impact on humans as well as on the environment. It has been proven to be toxic towards all forms of life and is difficult to transform or detoxify. Nitrilases are a practical and safe solution to such a problem as it does not require any assistance of environmentally harmful treatment process. Its effectiveness is further enhanced by the fact that it can remain active in the non-aqueous environment too. Upon transferring nitrilases, microorganisms have also shown tolerance toward nitriles waste, which further enhances their significance. Nitrilases and nitrilase-producing microbes have a great potential for remediating cyanide group containing wastes.
CONCLUSION
The current focus of academia and industry is to find ways to minimize environmental harm by utilizing green chemistry tools. Biocatalysis is one such tool that can effectively reduce hazardous chemical generation by reducing or even eliminating hazardous by-products. Furthermore, some important characteristics of biocatalysts, such as high selectivity and stereo-regio- specificity, removes the need for down processing of the product for stereo-isomeric conversion. Nitrilase is a group of enzymes that converts highly toxic cyanide and nitrile group into their respective carboxylic acid or amides without generating toxic by-products in the process. Some of the interesting characteristics of nitrilases, such as very high selectivity, adaptability, and tolerance to the organic solvent system, make them highly potential for biomedical and biotechnological application. A wide range of microorganisms have the capabilities to metabolize different types of nitrile compounds with varying degrees of hydrolysis. Nitrilase produced by such microorganisms has been recognized for the synthesis of several pharmaceutical and other fine chemical compounds. Many of the cyanide and nitrile conversion reactions which required environmentally hazardous processes, are now done with ease using nitrilase enzyme. The new and unique applications of nitrilase have been found recently in drug discovery, but investigations are required to identify further applications in the medical field. Cyanide detoxification could be one of the interesting aspects of it. Nitrilases are a new class of enzymes in comparison to several commercialized enzymes such as cellulases, pectinases, oxidoreductases, dehydrogenases, amylases, and lipases. So investigations are required to find more novel nitrilases from a wide variety of sources. One of the key limitations of nitrilases has been so far the high specificity of nitrilase which can be overcome by exploring nitrilases with broad substrate utilization capabilities. In particular, the use of nitrilase to synthesize optically pure chemical compounds from dinitriles, cyanohydrins, and other aromatic nitriles are worth exploring. Furthermore, to fully utilize the biotechnological potential of nitrilase, enzyme modification and reengineering need to be investigated. Databases of gene pools available as the open-source could also reveal novel nitrilases with unknown properties. The bioinformatics analysis work on nitrilases is much needed as more information regarding its 3D structure, specificity, and other biochemical properties can elucidate the role of such enzyme in nature. Recent advances on nitrilase have broadened the knowledge of nitrilases, and the utilization of genetic engineering tools has set future prospects bright.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


