All published articles of this journal are available on ScienceDirect.
Heterologous Expression of Transcription Factor AtWRKY57 Alleviates Salt Stress-Induced Oxidative Damage
Abstract
Background:
WRKY transcription factors play important roles in the responses to abiotic stresses, seed dormancy, seed germination, developmental processes, secondary metabolism, and senescence in plants. However, molecular mechanisms of WRKY transcription factors-related abiotic stress tolerance have not been fully understood.
Methods:
In this investigation, transcription factor AtWRKY57 was introduced into cell lines of rice (Oryza sativa L.), tobacco (Nicotiana tabacum), and white pine (Pinus strobes L.) for characterization of its function in salt stress tolerance. The purpose of this investigation is to examine the function of AtWRKY in a broad sample of plant species including monocotyledons, dicotyledons, and gymnosperms.
Results:
The experimental results demonstrated that heterologous expression of transcription factor AtWRKY57 improves salt stress tolerance by decreasing Thiobarbituric Acid Reactive Substance (TBARS), increasing Ascorbate Peroxidase (APOX) and Catalase (CAT) activity under salt stress. In rice, overexpression of transcription factor AtWRKY57 enhances expression of Ca2+-dependent protein kinase genes OsCPk6 and OsCPk19 to counteract salt stress.
Conclusion:
These results indicated that transcription factor AtWRKY57 might have practical application in genetic engineering of plant salt tolerance throughout the plant kingdom.
1. INTRODUCTION
High salinity is one of the main factors limiting plant growth and crop productivity. The responses of plants to salt stress are an important topic for the biotechnological application plant salt tolerance in the field of abiotic stress. Transcription Factors (TFs) ranging from bZIP, AP2/ERF, NAC, zinc finger proteins, and MYB proteins to WRKY highly influence the efficiency of salt stress-induced gene expression under salt stress. Overexpression of these transcription factors will provide new opportunities for the engineering of plant tolerance to salt stress [1]. In cotton, 42 relevant/related genes were induced by the NaCl treatment and they may be candidate genes as potential markers of tolerance to salt stress [2]. In wheat, transcription factor TaWRKY79 enhanced the level of tolerance to salinity stress via reducing the sensitivity to ABA in the TaWRKY79 transgenic plants, indicating that TaWRKY79 operates in an ABA-dependent pathway [3]. In Triticum aestivum cv. (Yuregir-89), Triticum turgidum cv. (Kiziltan-91), and Triticum monococcum (Siyez), expression analysis of transcription factors TaWLIP19, TaMBF1, TaWRKY10, TaMYB33 and TaNAC69 indicated that all five selected genes in Kiziltan-91 were induced, suggesting that transcription factors might be used for determination of salinity-tolerant for molecular breeding studies [4].
WRKY transcription factors are plant-specific, zinc finger-type transcription factors that are involved in abiotic stress tolerance in a large number of plant species. Analysis of DNA orthology and gene motif compositions indicated that WRKY members in many plant species generally shared the similar motifs. In Gossypium aridum, 28 salt-responsive GarWRKY genes were identified under stress. Overexpression of GarWRKY17 and GarWRKY104 in Arabidopsis demonstrated that these transcription factors would be potential candidates for the genetic improvement of cotton salt stress tolerance [5]. In peanut, transcription factors (NAC, bHLH, WRKY, AP2/ERF) are differentially expressed under salinity stress [6]. In cotton, silencing of C4 (encodes WRKY DNA-binding protein) can significantly enhance cotton susceptibility to salt stress [7]. In foxtail millet (Setaria italica), salt stress-induced methylation in WRKY transcription factor genes and modulates the expression of corresponding genes [8]. In Citrus, a total of 1831 differentially expressed genes were identified and a multitude of transcription factors including WRKY, NAC, MYB, AP2/ERF, bZIP, GATA, bHLH, ZFP, SPL, CBF, and CAMTA was related to cell wall loosening and was also involved in salt stress [9].
Transcription factors could differentially regulate salt stress tolerance. Basic region/leucine zipper (bZIP) transcription factors play key roles in plant growth, development, and stress signaling and perform as crucial regulators in ABA-mediated stress response in plants [10]. In ramie, a bZIP transcription factor BnbZIP2 may act as a positive regulator in response to high-salinity stress [11]. The homeodomain leucine zipper (HD-Zip) transcription factors modulate plant growth and response to environmental stresses. In Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays), poplar (Populus trichocarpa), soybean, and cucumber (Cucmis sativus), HD-Zip transcription factors have important roles in dehydration and salt stress [12]. Trihelix transcription factors, which are characterized by containing a conserved trihelix (helix-loop-helix-loop-helix) domain that binds to GT elements, play roles in abiotic stress responses by regulating the expression of stress tolerance genes that result in reduced reactive oxygen species and lipid peroxidation [13]. NAC (NAM ATAF CUC) transcription factors are involved in ethylene-modulated salt tolerance. In apple, MdNAC047 transcription factor was significantly induced by salt treatment and its overexpression conferred increased tolerance to salt stress [14]. WRKY transcription factors appear as important regulators of abiotic stresses tolerance. In tomato, SlWRKY3 binds to the consensus CGTTGACC/T W box and regulates expression of stress-related genes, indicating that SlWRKY3 is an important regulator of salinity tolerance [15]. In Reaumuria trigyna, WRKY transcription factor RtWRKY1 was induced by salt stress and ABA treatment. Overexpression of RtWRKY1 enhanced root length and fresh weight of the transgenic lines under salt stress [16]. In maize, expression of the ZmWRKY17 was up-regulated by salt and ZmWRKY17 acts as a negative regulator involved in the salt stress responses through ABA signaling [17].
NAC transcription factor proteins play important roles in salt stress responses. In Cucumis melo, CmNAC14 regulates expression of salt stress-related genes. Overexpression of CmNAC14 increased the sensitivity of transgenic lines to salt stress [18]. In Chickpea (Cicer arietinum L.), the transcript levels of CarNAC4 were enhanced in response to abiotic stresses. Over-expression of CarNAC4 improved tolerance to salt stresses [19]. In wheat, TaNAC29 was involved in response to salt. TaNAC29 confers salt stress tolerance through reducing H2O2 accumulation by enhancing the antioxidant system [20]. In tomato, SlNAC4-SlNAC10 genes are involved in the response to salt stress [21]. In Arabidopsis thaliana, AtNAC2 is a transcription factor that regulates salt stress and incorporates the environmental and endogenous stimuli into the process of plant lateral root development [22].
WRKY transcription factors play important regulatory roles in plant development and defense response. In Salvia miltiorrhiza, a total of 61 SmWRKYs were cloned [23]. In cucumber, a total of 55 WRKY genes were identified [24]. In Arabidopsis, the expression of AtWRKY25, AtWRKY26, and AtWRKY33 was related to NaCl, and osmotic stress [25]. In canola (Brassica napus L.), 46 WRKY genes were identified to be involved in stress [26]. In rice (Oryza sativa), OsWRKY genes contributing to salt stress tolerance-realted biological processes or signal transduction pathways [27]. In knapweed (Centaurea maculosa), differential gene expression was observed for a putative serpin (CmSER-1) and a calmodulin-like (CmCAL-1) protein. In dandelion (Taraxacum officinale), differential gene expression was observed for a putative protein phosphatase 2C (ToPP2C-1) and cytochrome P-450 (ToCYP-1) protein. These genes are involved in plant stress responses [28]. In Arabidopsis, 49 of the 72 AtWRKY genes were differentially regulated in the plants infected by an avirulent strain of the bacterial pathogen Pseudomonas syringae [29].
Transcription factor WRKY57 is known to function in adaptation to abiotic stresses. In banana (Musa acuminate), WRKY57 is necessary for developing drought-resilient plants [30]. In Arabidopsis, WRKY57 directly binds to the promoters of JASMONATE ZIM-DOMAIN1 (JAZ1) and interacts with nuclear-encoded SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 to allow fine regulation of defense [31]. Overexpression of the Arabidopsis WRKY57 transcription factor AtWRKY57 in rice improved not only drought tolerance but also salt and PEG tolerance, indicating its potential role in crop improvement [32]. In grape (Vitis vinifera L.), the WRKY57-like transcription factor has an important role in some biological processes including cell rescue, protein fate, secondary metabolism, and regulation of transcription [33]. Leaf senescence is regulated by diverse developmental and environmental factors. WRKY57 interacts with the AUX/IAA protein IAA29 to regulate leaf senescence process as a common component of the JA- and auxin-mediated signaling pathways [34]. Constitutive expression of WRKY57 conferred drought tolerance by regulating expression of stress-responsive genes (RD29A, NCED3, and ABA3). ChIP assays demonstrated that WRKY57 could directly bind the W-box of RD29A and NCED3 promoter sequences. Increased WRKY57 expression enhanced drought stress tolerance by increasing ABA levels that will increase drought stress tolerance in plants [35]. In this investigation, we demonstrated that transcription factor AtWRKY57 might have practical application in genetic engineering of plants for salt stress tolerance throughout the plant kingdom.
2. MATERIALS AND METHODS
2.1. Plasmid Constructs
The expression vector pBI121 and the cDNA of WRKY57 were used to construct the pBI-WRKY57 expression vector. Restrict enzymes Kpn I and Xba I (Promega, Madison, WI, USA) were used to digest both the pBI121 vector and the WRKY57 cDNA at 37oC, DNA bands of the pBI121 vector and the WRKY57 cDNA were purified using QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA, USA). The 864-bp fragment of the WRKY57 cDNA was ligated with the DNA of digested pBI121 [36] to produce the expression vector pBI-WRKY57. Vector pBI-WRKY57 was introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation.
2.2. Agrobacterium-Mediated Transformation
Transgenic cell lines of rice (Oryza sativa L.), tobacco (Nicotiana tabacum), and white pine (Pinus strobes L.) were prepared as previously described [37, 38], using Agrobacterium tumefaciens strain LBA4404 carrying pBI-WRKY57 to transform cell cultures. Transgenic cells of rice, tobacco, and white pine were sub-cultured on a liquid proliferation medium for six weeks to obtain large quantities of cell cultures for further analysis. After 6 weeks, 50-70 mg of tissue/l cultures can be produced each week, and they were then used molecular analysis including PCR, Southern blot analysis, and northern blotting analysis.
2.3. Polymerase Chain Reaction and Southern Blot Analyses of Transgenic Cultures
Polymerase Chain Reaction (PCR) and Southern blotting analysis of putative transgenic cell lines of rice, tobacco, and white pine were conducted as previously described [37, 38]. 500 mg fresh cells of rice, tobacco, and white pine control and putative transgenic cell lines were used to isolate genomic DNA, using a Genomic DNA Isolation Kit (Sigma) following the manufacturer’s protocol. PCR was done in a PTC-100TM Programmable Thermal Controller (MJ Research, San Francisco, CA, USA). The primers used are the nptII forward primer (nfp) 50-ACAAACAGACAATCGGCTGC-30 and the reverse primer (nrp) 50-AAGAACTCGTCAAGAAGGCG-30, as well as the transcription factor WRKY57 forward primer (zfp) 5’-ATGAACGATCCTGATAATCCCGATC-3’ the reverse primer (zrp) 5’-TCAAGGGTTGCGCATAGTTTGAGG-3’ were used. Southern blot analysis was conducted as previously described [38]. 25 μg DNA was digested overnight at 37 oC. Probes (864 pb fragment of WRKY57) were labeled by Digoxigenin (DIG) (Roche Diagnostics, Indianapolis, IN, USA).
2.4. RNA Isolation and Northern Blot Analysis
Total RNA was isolated from 5 g fresh cell cultures of rice, tobacco, and white pine control and putative transgenic cell lines, respectively, using a RNeasy Mini Plant Kit (Germantown, MD, USA) following the manufacturer’s protocol. Then, 6 μg RNA from rice, tobacco, and white pine transgenic cells were separated by agarose-gel electrophoresis. Electrophoresis and northern blotting of RNAs were performed as previously described [36]. Digoxigenin (DIG)-labelling WRKY57 DNA (864 pb) (Roche Diagnostics) was used as a hybridization probe. Equal loading of RNA samples of rice, tobacco, and white pine was verified on the control of tobacco 25SrRNA. After PCR, Southern blotting and northern blotting analyses, four cell lines each containing one copy of the pBI-WRKY57 T-DNA were selected from rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, andNt6), and white pine (Ps1, Ps2, Ps3, and Ps4) and used for salt-induced oxidative damage experiments.
2.5. Salt Treatment of Transgenic Cell Lines
Salt treatment was applied by adding different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM) to the media used for transgenic cell cultures, which consisted of TE medium [38] containing 0.5 μM indole-3-butyric acid, 8.9 μM BA. The effects of NaCl on growth of cell cultures of rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, andNt6), and white pine (Ps1, Ps2, Ps3, and Ps4) were examined by culture of cell cultures on growth medium containing different concentrations of NaCl, as previously described [36-38]. The average growth was expressed as mg/g FW/day.
2.6. Thiobarbituric Acid Reactive Substances (TBARS) Determination
The amount of thiobarbituric acid reactive substances (TBARS) was determined using the method of thiobarbituric acid (TBA) reaction as described previously [38]. Cell cultures (1 g) of rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, andNt6), and white pine (Ps1, Ps2, Ps3, and Ps4) were homogenized in 3 ml of 20% (w/v) trichloroacetic acid (TCA). The homogenate was centrifuged at 5,000 rpm for 20 min and mixed with 20% TCA containing 0.5% (w/v) TBA and100 ll 4% BHT in ethanol at 1:1. The amount of thiobarbituric acid reactive substances was determined using the method previously described [36].
2.7. Antioxidant Enzymes Ascorbate Peroxidase (APOX) and Catalase (CAT) Activity Determination
Determination of activities of APOX and CAT were determined as described previously [36-38]. Two grams of non-transgenic control and transgenic cell cultures of rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, andNt6), and white pine (Ps1, Ps2, Ps3, and Ps4) were homogenized under ice-cold conditions in 3 ml of extraction buffer, containing 50 mM phosphate buffer (pH 7.4), 1 mM EDTA, 1 g PVP, and 0.5% (v/v) Triton X-100 at 4 oC [36-38]. The homogenates were centrifuged at 10,000 × g for 20 min and the supernatant fraction was used for the assays. APOX activity was measured immediately in fresh extracts and was assayed as described [36]. CAT activity was detected in a 3 ml 50 mMpotassium phosphate buffer (pH 7.8) containing 3 mMH2O2, as previously described [36].
2.8. Expression Analysis of OsCPK Genes
Expression of OsCPK6 and OsCPK19 was analyzed by northern blotting by the method described previously [37, 38]. Twenty micrograms of total RNA of rice (Os1, Os2, Os3, and Os4) was used. The PCR-amplified fragments of OsCPK6 (amplified by forward primer 5’- atgggcaactactactcgtg -3’ and reverse primer 5’- acgtacaggttgtcctcgt -3’) and OsCPK19 (amplified by forward primer 5’- ggagcaaacggttatggcta -3’ and reverse primer 5’- gcgcttagagatggatttgc -3’) were labeled by Digoxigenin (DIG) (Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, IN, USA) and were used as a hybridization probe [37, 38].
2.9. Statistical Analyses
Data on growth rate, amount of TBARS, APOX and CAT activity, amount of OsCPK6 and OsCPK19 obtained from different experiments were analyzed in Graphpad Prism 6 software. The significant differences between mean values derived from three independent biological experiments were determined using the least significant difference test at 5% level of probability.
3. RESULTS
3.1. Molecular Analyses of Transgenic Cell Lines
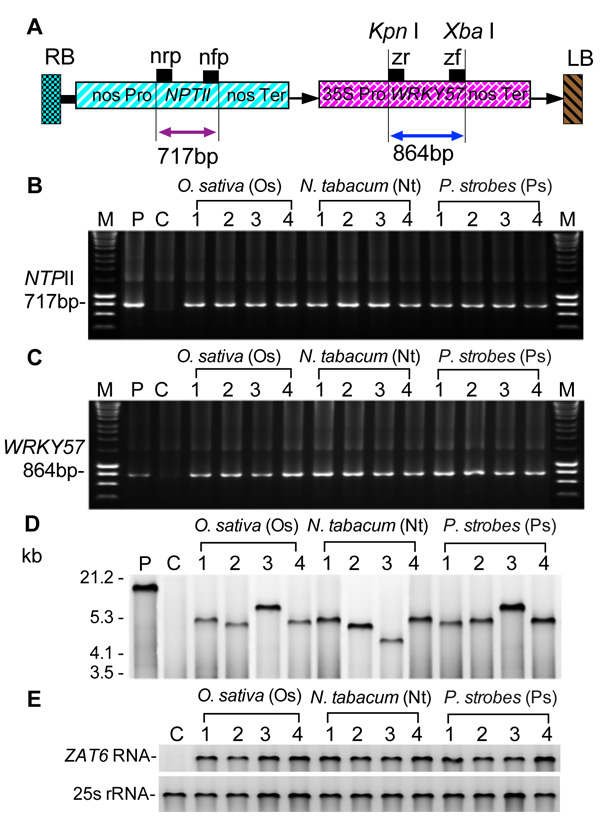
Transgenic cell lines of rice (Oryza sativa L.), tobacco (Nicotiana tabacum), and white pine (Pinus strobes L.) were obtained through Agrobacterium tumefaciens-mediated transformation and were examined by PCR, Southern blotting, and northern blotting (Fig. 1). A total of 109 transgenic cell lines including 31 cell line from rice, 36 cell line from tobacco, and 42 cell line from white pine, were obtained after PCR analysis using primers for neomycin phosphotransferase II gene and primer for WRKY57. After confirmation by PCR, four cell lines were randomly selected and used for further analysis. The presence of the 717-bp band amplified by primers nrp and nfp from templates obtained from transgenic cells and absence of the 717-bp band amplified by primers nrp and nfp indicate that the T-DNA (Fig. 1A) is integrated into the genome. This is further confirmed by PCR using primers for WRKy57 that the 864-bp band amplified by primers zf and zr from templates obtained from transgenic cells and absence of the 864-bp band amplified by primers zf and zr (Fig. 1B). Integration of T-DNA into the genome of rice, tobacco, and white pine was also confirmed by Southern and northern blotting analisys (Figs. 1C, D).
3.2. Growth of Cell Lines Under Different Concentrations of NaCL
Growth rate (Fig. 2) of transgenic cell lines of rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, and Nt4), and white pine (Ps1, Ps2, Ps3, and Ps4) were measured 3 days after transgenic cell cultures were transferred into media containing different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM). The medium containing no NaCl was used as the control of salt stress. A significant difference in growth rate was obtained when transgenic cell cultures were cultured on media containing 150, 200, 250, or 300 mM NaCl, compared to the control. Similar results were obtained in all 12 transgenic cell lines derived from rice, tobacco, and white pine, indicating that overexpression of WRKY57 enhances salt stress tolerance in plants including monocotyledons (rice, Fig. 2A), dicotyledons (tobacco, Fig. 2B), and gymnosperms (white pine, Fig. 2C).

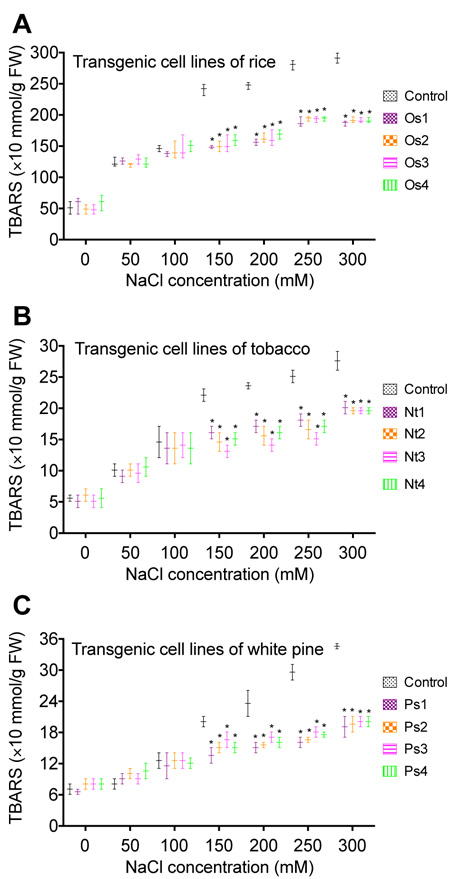
3.3. Thiobarbituric Acid Reactive Substance (TBARS) Changes Under Different Concentrations of NaCl
The salt stress increased the formation of lipid peroxidation via increasing the rate of Reactive Oxygen Species (ROS) formation. Decreasing lipid peroxidation via genetic engineering approaches in cells will increases cells tolerance to salt stress. To determine if salt stress tolerance enhanced by overexpression of WRKY57 is related to the change of Thiobarbituric Acid Reactive Substance (TBARS), TBARS was measured in transgenic cell lines of rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, and Nt4), and white pine (Ps1, Ps2, Ps3, and Ps4) 3 days after cell cultures were transferred into media containing different concentrations of NaCl (Fig. 3). A significant decrease in the amount of TBARS was obtained in rice (Fig. 3A), tobacco (Fig. 3B), and white pine (Fig. 3C) when transgenic cell cultures were cultured on media containing 150, 200, 250, or 300 mM NaCl, compared to the control. Similar results were obtained in all 12 transgenic cell lines derived from rice, tobacco, and white pine, indicating that overexpression of WRKY57 enhances salt stress tolerance by decreasing the amount of TBARS. Compared to the control, no significant decrease in the amount of TBARS was obtained when 50 mM and 100 mM NaCl were applied to the transgenic cells.
3.4. Effect of WRKY57 Overexpression on APOX and CAT Activity
To examine if salt stress tolerance enhanced by overexpression of WRKY57 is involved in the change of antioxidant enzymes Ascorbate Peroxidase (APOX) and Catalase (CAT) activity, APOX and CAT activity were measured in transgenic cell lines of rice (Os1, Os2, Os3, and Os4), tobacco (Nt1, Nt2, Nt3, and Nt4), and white pine (Ps1, Ps2, Ps3, and Ps4) 3 days after cell cultures were transferred into media containing different concentrations of NaCl (Fig. 4A). Increased APOX and CAT activity were observed in transgenic cell lines of rice (Figs. 4A, D), tobacco (Figs. 4E, 4F) and white pine (Figs. 4C, F) indicating that overexpression of WRKY57 enhances salt stress tolerance by increasing the activity of APOX and CAT. Compared to the control, no significant increase in APOX and CAT activity was obtained when 50 mM and 100 mM NaCl were applied to the transgenic cells.

3.5. Influence of NaCl on Expression of Rice Ca2+-Dependent Protein Kinase Gene Oscpk6 and Oscpk19
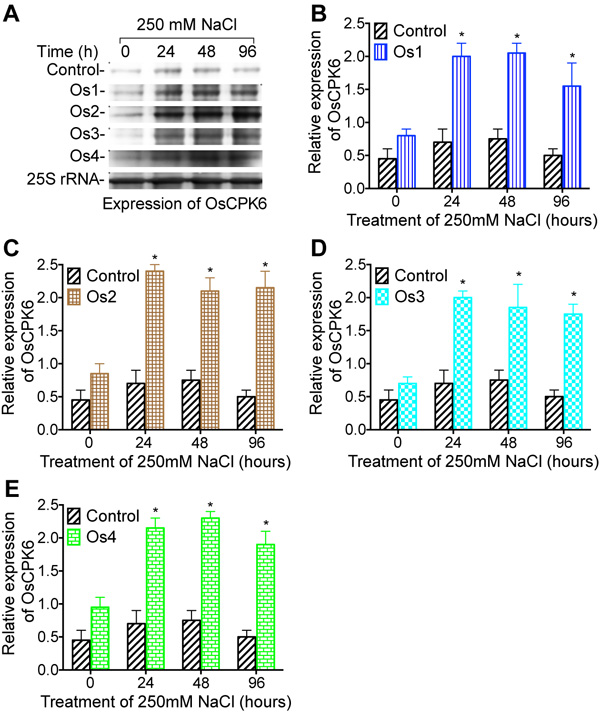
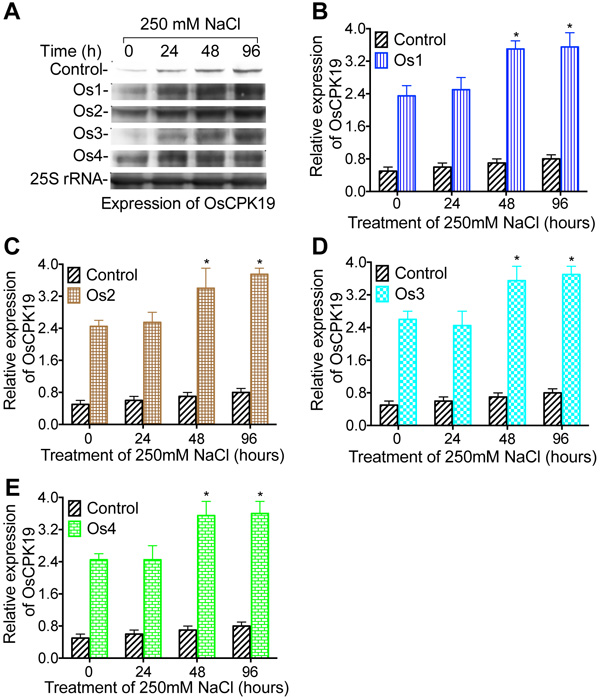
To investigate if salt stress tolerance enhanced by overexpression of WRKY57 is related to the expression of Ca2+-dependent protein kinase gene OsCPK6, the amount of OsCPK6 mRNA was measured by northern blotting in transgenic cell lines of rice (Os1, Os2, Os3, and Os4), 24, 48, and 96 hours after cell cultures were transferred into media containing 250 mM NaCl (Fig. 5). Increase in the amount of OsCPK6 mRNA was obtained in rice 24, 48, and 96 hours after transgenic cell cultures were treated by 250 mM NaCl in all four rice cell lines Quantitative analysis of expression of Ca2+- dependent protein kinase gene OsCPK9 demonstrated that the amount of OsCPK6 mRNA was increased significantly (Figs. 5B, E), compared to the control. Northern blotting analysis of expression of Ca2+- dependent protein kinase gene OsCPK19 (Fig. 6) in WRKY57 transgenic cell lines Os1, Os2, Os3, and Os4 demonstrated that the amount of OsCPK19 mRNA was increased under 250 mM NaCl treatment at 24, 48, and 96 h. Quantitative analysis of the expression of Ca2+- dependent protein kinase gene OsCPK9 demonstrated that the amount of OsCPK19 mRNA was increased significantly (Figs. 6B, E).
4. DISCUSSION
The WRKY transcription factors play very important roles in plant growth, some development stages, and plants responses to abiotic stresses. In Sesame (Sesamum indicum L.), manipulating WRKYs could improve resistance to waterlogging stress and drought stress [39]. In Arabidopsis, WRKY75 functions as a new component of the GA-mediated signaling pathway to positively regulate flowering in Arabidopsis [40]. WRKY71 activity hastens flowering by providing a means for the plant to complete its life cycle in the presence of salt stress [41]. WRKY36 is a negative regulator of HY5 and UVB represses WRKY36 via UVR8 to promote the transcription of HY5 and photomorphogenesis [42]. In oilseed rape (Brassica napus), BnWRKY15 overexpression simultaneously increased the susceptibility of B. napus to S. sclerotiorum and down-regulated BnWRKY33 after different durations of infection [43]. AtWRKY11 and AtWRKY17 are not strictly limited to plant defense responses but are also involved in conferring stress tolerance [44]. Overexpression of WRKY27 displays significantly decreased pollen viability ans is involved in proper plant biomass accumulation and male fertility [45]. In Arabidopsis, WRKY43 acts as a potent modulator of fatty acid desaturation and seed filling, which results in increased tolerance to abiotic stress [46]. WRKY46, WRKY54, and WRKY70, are involved in both BR-regulated plant growth and drought response [47]. Transcription factor WRKY57 enhances salt stress tolerances in plant [32] and is important for developing drought-resilient crops [30]. In grape (Vitis vinifera L.), the WRKY57-like protein was strongly upregulated by UV-C irradiation and have great implications for further studies [33-35].
Environmental stress can affect the viability of an organism and may affect the genome or the proteome by contributing to protein damage or misfolding [48-52]. Plant responses to abiotic stress require two protective measures, one is to reduce stress-inflicted damage to cellular structures, another is the molecular mechanism that efficiently removes damaged and toxic macromolecules [53]. The antioxidant enzyme activities (Superoxide Dismutase (SOD), Ascorbate Peroxidase (APX), and Glutathione Reductase (GR)) and the contents of malondialdehyde (MDA) and H2O2 are important in reactive oxygen scavenging systems that could enhance salinity tolerance and alleviated salinity-induced damage in plants [54]. The Ca2+- dependent protein kinase gene OsCPK4 gene is a member of the complex gene family of calcium-dependent protein kinases in rice (Oryza sativa). OsCPK4 is a positive regulator of the salt and drought stress responses in rice that protect cellular membranes and cell signaling transduction from stress-induced oxidative damage [55].
Inorganic Nanoparticles (NP) have been used in different applications and recent research demonstrated that NP cause adverse effects via induction of an oxidative stress. In vitro methods using oxidative stress biomarkers to measure stress were designed and standardized for conventional NP [56]. It has been reported that a melon Superoxide Dismutase (SODB) is involved in oxidative stress. SODB could exert its antioxidant properties by inducing the endogenous antioxidant defense [57]. OsCEST and AtCEST were mainly transcribed in photosynthetic tissues. Overexpression of CEST enhanced tolerance not only to salt stress but also to drought stress, and high-temperature stress, which causes photooxidative stress [58]. In potato (Solanum tuberosum L. cv. Taedong Valley), a higher level of GSH homeostasis and higher glyoxalase activity inhibit the accumulation of methylglyoxal under salt stress [59]. Investigation of exogenous selenium (Se) in the antioxidant defense demonstrated that exogenous selenium improves stress tolerant to salt stress-induced oxidative damage and enhances their antioxidant defense and MG detoxification systems [60].
Salinity stress-induced production of Reactive Oxygen Species (ROS) and associated oxidative damage is one of the major factors limiting crop production in saline soils [61]. Superoxide Dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and glutathione reductase in chickpea plants were up-regulated by salt stress [52]. Transcription factors, such as MYB, WRKY, and zinc finger transcription factors may be regulated by environmental stresses, such as hormones, drought, cold, heat, and pathogens as well as by Pi starvation [62-64]. In tomato, high-antioxidant enzyme activities were induced during the response to salt stress [63]. Intricate signaling network responding to the diverse environmental stress could be developed by plants. The transcript expression of SA biosynthetic gene ICS1 and CAT and SOD antioxidative enzymes demonstrated upregulation during stress, indicating biochemical and molecular pathways to maneuver salt stress tolerance of the transgenic plants [64-70].
CONCLUSION
In this investigation, transcription factor AtWRKY57 was introduced into cell lines of rice (Oryza sativa L.), tobacco (Nicotiana tabacum), and white pine (Pinus strobes L.) for characterization of its function in salt stress tolerance. The purpose of this investigation is to examine the function of AtWRKY throughout the plant kingdom including monocotyledons, dicotyledons, and gymnosperms. The experimental results demonstrated that heterologous expression of transcription factor AtWRKY57 improves salt stress tolerance by decreasing Thiobarbituric Acid Reactive Substance (TBARS), increasing Ascorbate Peroxidase (APOX) and Catalase (CAT) activity under salt stress. In rice, overexpression of transcription factor AtWRKY57 enhances expression of Ca2+-dependent protein kinase genes OsCPk6 and OsCPk19 to counteract salt stress. These results indicated that transcription factor AtWRKY57 might have practical application in genetic engineering of plant salt tolerance throughout the plant kingdom.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The author is grateful to Wells Thompson and Jennifer Whitley for their critical reading of the manuscript. The author appreciates Nicole Franco-Zorrilla, Tinya Backiyarani, and Ambrosia Oluwadahunsi for their support in preparing transgenic cell cultures. This work was supported by a grant from the Education Committee of Hubei Providence of China (Grant No. D20101306).


