All published articles of this journal are available on ScienceDirect.
Optimization of Lipase Production in Solid-State Fermentation by Rhizopus Arrhizus in Nutrient Medium Containing Agroindustrial Wastes
Abstract
Background:
Rhizopus arrhizus is a potential microorganism for lipase production. Solid-state fermentation is used for microbial biosynthesis of enzymes, due to advantages, such as high productivity, utilization of abundant and low-cost raw materials, and production of enzymes with different catalytic properties.
Objective:
The objective of the research is optimization of the conditions for lipase production in solid-state fermentation by Rhizopus arrhizus in a nutrient medium, containing agroindustrial wastes.
Method:
Biosynthesis of lipase in solid-state fermentation by Rhizopus arrhizus was investigated. The effect of different solid substrates, additional carbon and nitrogen source, particles size and moisture content of the medium on enzyme production was studied. Response surface methodology was applied for determination of the optimal values of moisture content and tryptone concentration. A procedure for efficient lipase extraction from the fermented solids was developed.
Results:
Highest lipase activity was achieved when wheat bran was used as a solid substrate. The addition of 1% (w/w) glucose and 5% (w/w) tryptone to the solid medium significantly increased lipase activity. The structure of the solid medium including particles size and moisture content significantly influenced lipase production. A mathematical model for the effect of moisture content and tryptone concentration on lipase activity was developed. Highest enzyme activity was achieved at 66% moisture and 5% (w/w) tryptone. The addition of the non-ionic surfactant Disponyl NP 3070 in the eluent for enzyme extraction from the fermented solids increased lipase activity about three folds.
Conclusion:
After optimization of the solid-state fermentation the achieved 1021.80 U/g lipase activity from Rhizopus arrhizus was higher and comparable with the activity of lipases, produced by other fungal strains. The optimization of the conditions and the use of low cost components in solid-state fermentation makes the process economicaly effective for production of lipase from the investigated strain Rhizopus arrhizus.
1. INTRODUCTION
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) are one of the most important classes of industrial enzymes. They take a second place after proteases and carbohydrases in world enzyme market and have a share of about 5% [1]. Lipases hydrolyse triglycerides into diglycerides, monoglycerides, glycerol and fatty acids. They are able to catalyse not only hydrolysis but also synthesis reaction (esterification and transesterification) in media with low water content [2].
Enzymes of industrial interest traditionally have been produced by Submerged Fermentation (SmF). Solid-State Fermentation (SSF) is defined as the fermentation process on moist solid substrate in the absence or near absence of free water [3]. However, SSF was used for enzymes production, due to its advantages, such as high productivity, generation of high-quality products, and use of abundant and low-cost raw materials such as agro industrial wastes. The use of complex matrix of agro industrial wastes as a culture medium also induces the production of different hydrolytic enzymes such as amylases and proteases in the same fermentation batch, or even pools of lipases with different catalytic properties [4]. Diaz et al. found that lipase produced by SSF of Rhizopus homothallicus was significantly thermostable, and the optimal operating temperature was 10°C higher in comparison to the enzyme produced in SmF [5].
SSF encounters problems related to mass and heat transfer phenomena associated with solid substrates. The use of natural solid substrates can hinder downstream processes, especially when extracting lipophilic enzymes such as lipases. Fermented solids have been used as naturally immobilized biocatalysts for synthesis reactions in lyophilized or dried form. This approach can lead to lower costs of enzyme preparations since no extraction and purification steps are carried out [3].
For lipase production in SSF by fungi strains, various wastes from the agricultural industry are used. Silva et al. used babassu cake with 6.25% molasses in the nutrient medium to produce lipase from Penicillium simplicissimum at 65% moisture [4]. Awan et al. used almond meal examining the effect of the additional carbon source and the mineral composition of the medium on lipase biosynthesis from Rhizopus oligosporus [6].
For optimization of the composition of the nutrient medium and cultivation conditions, many authors used mathematical experimental designs. Fallony et al. conducted an experimental design and developed a mathematical model to optimize the composition of the nutrient medium and the conditions for SSF [7]. A similar study was conducted by Godoy et al. which by the Plackett-Burman design found the optimal values of several factors influencing lipase biosynthesis in SSF of Penicilium simplicissimum - particle size, initial moisture, initial pH, inoculum concentration and molasses concentration [8].
An important step in lipase production by SSF is the development of methodology for enzyme extraction. Silva et al. carrying out an experimental design found that the significant factors influencing lipase extraction obtained in SSF of Penicillium simplicissimum were pH, Tween 80 concentration, temperature and buffer molarity [4].
The aim of the research is optimization of the conditions for lipase production in SSF by R. arrhizus in a nutrient medium, containing agroindustrial wastes as solid substrates.
2. MATERIAL AND METHODS
2.1. Microorganism
The studied Rhizopus arrhizus strain used in this study was provided by Biovet® Peshtera. It was grown in the following medium, g/l: malt extract 10.00; yeast extract 4.00; glucose 4.00; agar-agar 20.00. pH was adjusted to 7.0. The strain was cultivated at 28°С for 14 days and stored at 4°С.
2.2. SSF and Media Preparation
SSF was carried out in 500 ml Erlenmeyer flasks. The flasks contained minimal nutrient medium of 10 g solid substrate and salt solution (g/l): NH4H2PO4 6.5, (NH4)2C2O4 0.90, MgSO4 0.95, KCl 0.95. pH of the salt solution was adjusted to 7.0, and the final moisture content was adjusted to the required value with the salt solution before autoclaving. After sterilization at 121°С for 30 min, the flasks were inoculated with 5 ml inoculum with 107 spores/ml and incubated at 30°C for 168 h.
2.2.1. Solid Substrates for Lipase Biosynthesis in SSF
The following solid substrates were studied for lipase biosynthesis: corn flour, wheat bran, rice bran, wheat flour, sunflower meal, sunflower cake, oat bran. SSF was performed with each of the substrates as described above.
2.2.2. Effect of Additional Carbon Source on Lipase Biosynthesis in SSF
Glucose, sucrose, starch and olive oil were tested as additional carbon sources for lipase biosynthesis. The solid substrate was supplemented with 1% (w/w) additional carbon source and SSF was performed as described above. The effect of glucose concentration on lipase production was studied in the range of 1-5% (w/w).
2.2.3. Effect of Additional Organic Nitrogen Source on Lipase Biosynthesis in SSF
The effect of the additional organic nitrogen source on lipase biosynthesis was studied by addition of 1% (w/w) nitrogen sources such as peptone, tryptone, yeast extract, soybean meal, cotton flour and fish meal. The effect of tryptone concentration was investigated in the range of 1-10% (w/w).
2.3. Optimization of Lipase Production in SSF by Response Surface Methodology
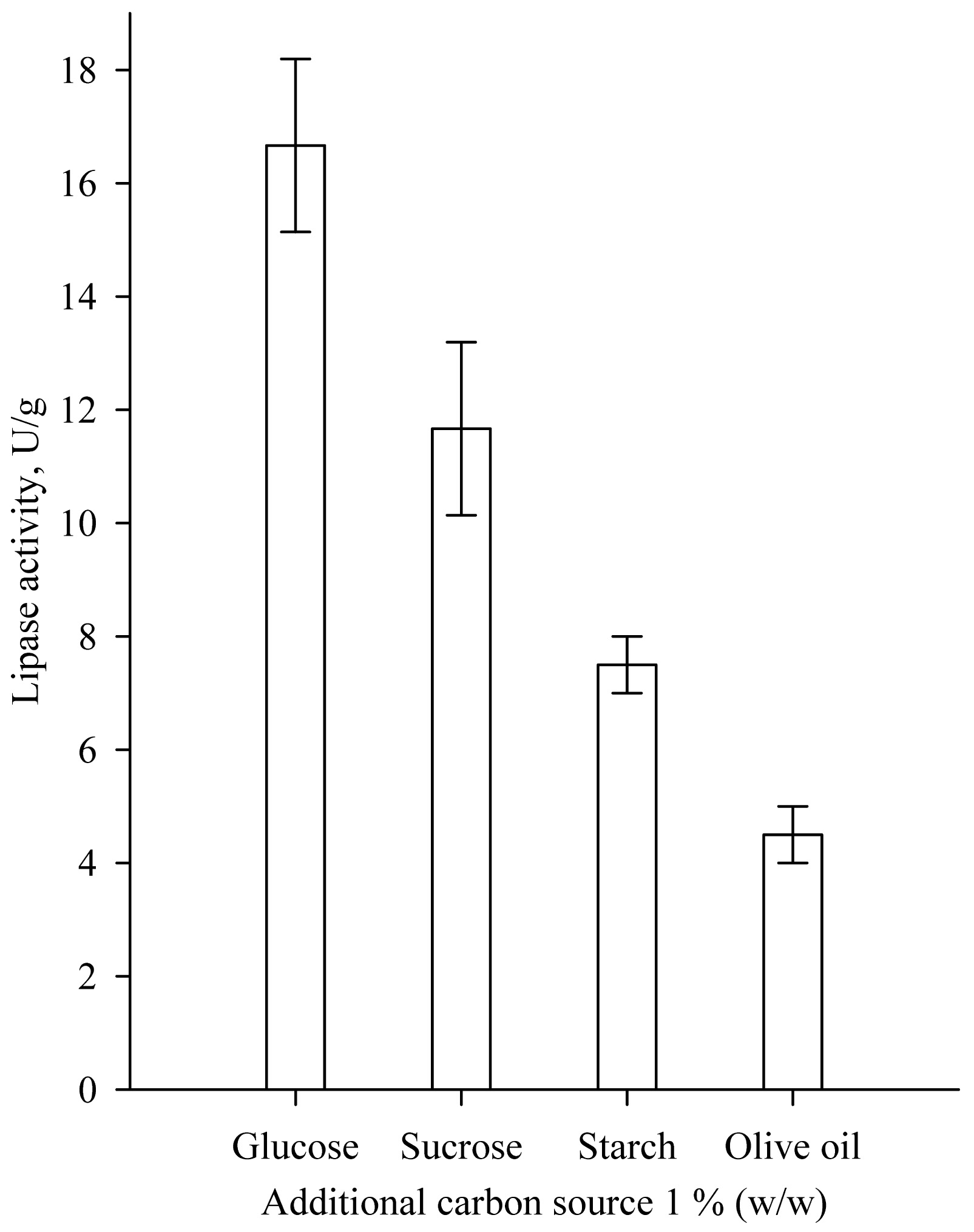
Response surface methodology and 22 Optimal Composite Design (OCD) with “star points” around the center point was used to determine the optimum concentrations of moisture and tryptone for lipase biosynthesis. The distance from the design center to a factorial point was a = ±1 [9, 10]. The quadratic regression model was expressed as follows:
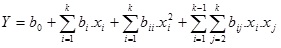 |
(1) |
where Y is the response variable, b the regression coefficients of the model and x the coded levels of the independent variables.
SygmaPlot software from Systat Software, Inc. was used for regression and graphical analysis. The independent variables participating in the 22 OCD and their values are presented in Table 1.
| Independent variable | -1 | 0 | +1 |
| Moisture (x1), %(w/w) | 50 | 60 | 70 |
| Tryptone (x2), %(w/w) | 3 | 5 | 7 |
2.4. Extraction of Lipase from the Fermented Solids
Following SSF, extraction of lipase from the fermented solids was performed with 50 ml eluent 1% Tween 80 for 30 minutes with constant agitation. Different salts and commercial surfactants in concentration of 0.5 and 1.0% were also tested as eluents. The solids were removed by filtration, the filtrates were centrifuged and tested for lipase activity. The results are presented in units lipase activity obtained from 1g of solid substrate.
2.5. Determination of Lipase Activity
For lipase activity determination the method described by Babu et al. [2], Saifuddin et al. [11], and Dobrev et al. [12] was adapted. The substrate was 30 mg ρ-nitrophenyl palmitate solution in 10 ml isopropanol, 90ml 0.05M Tris-HCl buffer with pH 7.2, 0.4g Triton X-100 and 0.1g gum Arabic. A reaction mixture consisting of 2.4 ml of the substrate solution and 0.1ml enzyme was incubated at 35°С for 30 min. The enzyme reaction was stopped with 1.0 mL 0.5 M EDTA with pH 8.0 and the absorbance at 405 nm was measured. One unit (U) of lipase activity was defined as the amount of enzyme forming 1 µmol ρ-nitrophenol for 1 min at 35°С and pH 7.2.
3. RESULTS AND DISCUSSION
3.1. Effect of Solid Substrate on Lipase Biosynthesis in SSF
The choice of solid substrate is of great importance to SSF, because in addition to being a source of nutrients, it forms the structure of the culture medium and determines the possibility of mass and heat transfer. The results for lipase biosynthesis on different solid substrates are shown in Fig. (1).
Highest lipase activity was noticed when wheat bran was used as a solid substrate. Significant lipase activity was also achieved with the use of other wastes from the agro and food industry, such as sunflower meal and sunflower cake, as well as oat bran and rice bran. The low price and availability of wheat bran make them a preferred solid substrate for carrying SSF [15].
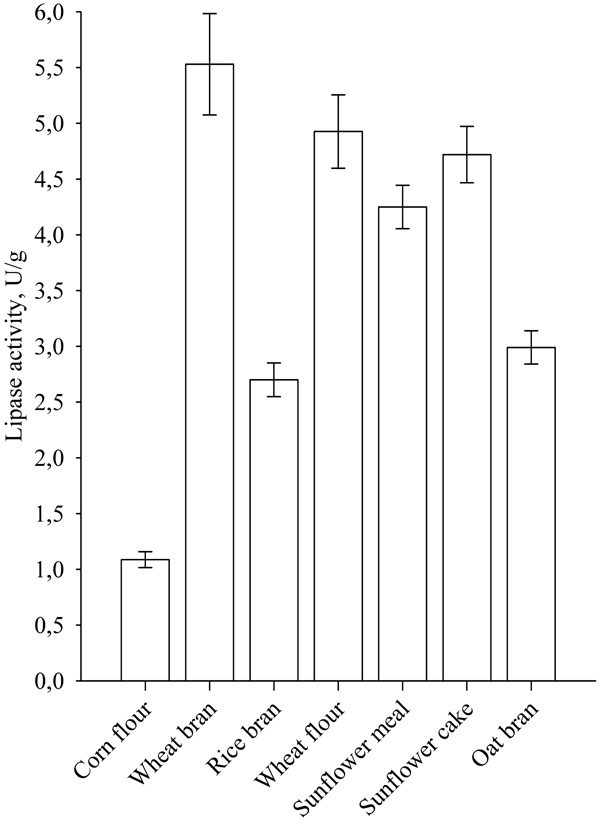
The effect of fermentation time on lipase biosynthesis, when wheat bran was used as solid substrate is presented in Fig. (2).
After 3 days of cultivation, a sharp peak of lipase activity was observed, with maximal activity being achieved at 168h, after which the activity was retained.
3.2. Effect of Additional Carbon Source on Lipase Biosynthesis in SSF
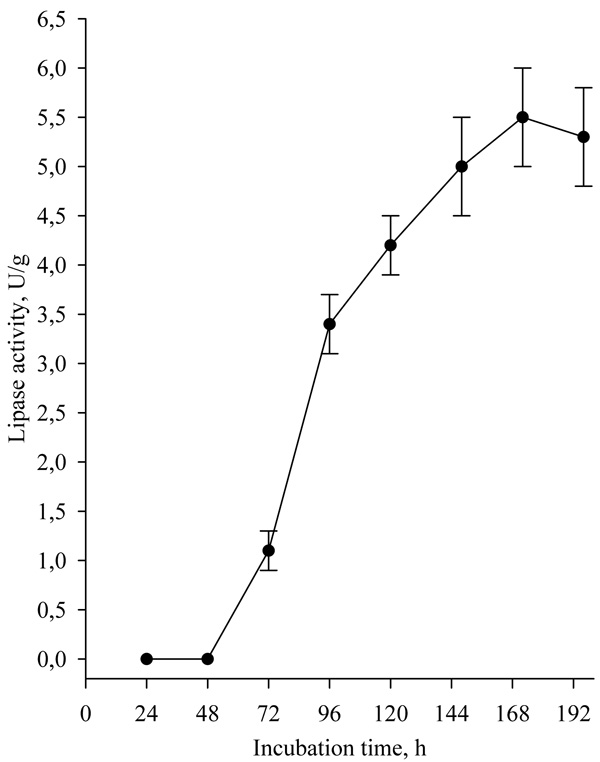
The effect of the additional carbon source to the medium of wheat bran on lipase biosynthesis is presented in Fig. (3). Significantly higher lipase activity was achieved by the use of glucose as an additional carbon source at a concentration of 1% (w/w) compared to the addition of starch and olive oil. The results obtained differ from the results of Awan et al. that found that the addition of olive oil and Tween 80 significantly increased lipase production by Rhizopus oligosporus compared to glucose addition [6].
For explanation of the results the chemical composition of the solid substrate wheat bran was determined (Table 2).
| Solid Substrate | Moisture, % |
Protein,
% |
Total Sugars, % |
Starch,
% |
Fat,
% |
Ash,
% |
|---|---|---|---|---|---|---|
| Wheat Bran* | 12.22±0.23 | 15.02±0.18 | 78.45±1.8 | 25.03±0.56 | 2.48±0.08 | 3.82±0.06 |
The chemical composition of wheat bran varies significantly according to the milling process and the nutritional components are typically in wide range. Proteins content is about 9.6-18.6%, total carbohydrate content is in the range of 60.0-75.0%, and starch varies in the range of 9.10-38.9% of dry matter [16]. The starch content of the used wheat bran was significantly high (25.03%). It should be noted that the use of starch as an additional carbon source in the nutrient medium significantly reduced lipase activity. This was probably due to the fine particle size and the high starch water holding capacity, which created problems in mass transfer and hindered strain development.
The influence of glucose concentration on lipase activity is presented in Fig. (4).
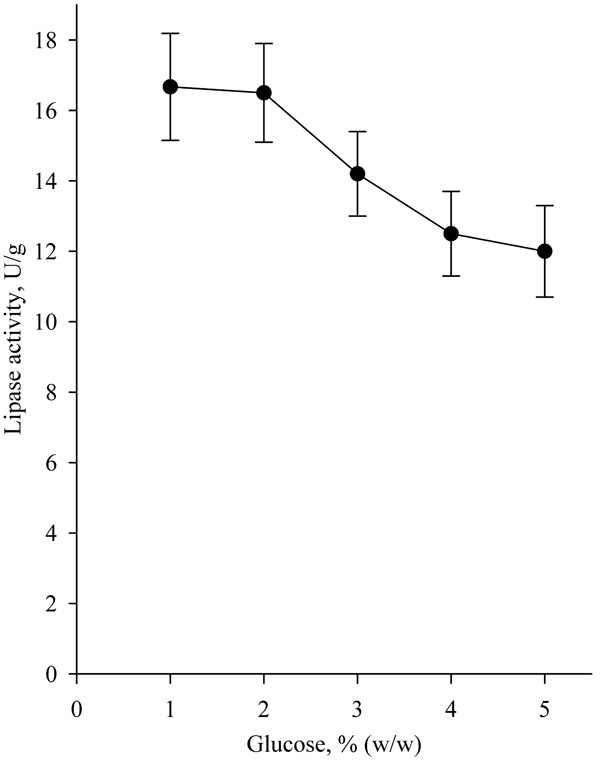
The results showed that the addition of glucose in concentrations higher than 2% (w/w) reduced lipase activity.
3.3. Effect of Additional Organic Nitrogen Source on Lipase Biosynthesis
The addition of 2% (w/w) tryptone, peptone and yeast extract as an additional source of organic nitrogen significantly increased lipase production (Fig. 5). Highest lipase activity was achieved when tryptone was used as an additional organic nitrogen source. The use of soybean meal, cotton flour and fishmeal led to a decrease in lipase activity. Similar results were obtained by Sun et al. for the supplementation of peptone to wheat bran and wheat flour solid substrates for SSF production of lipase by Rhizopus chinensis. The authors reported also an increase in lipase activity with the addition of yeast and beef extract [17].
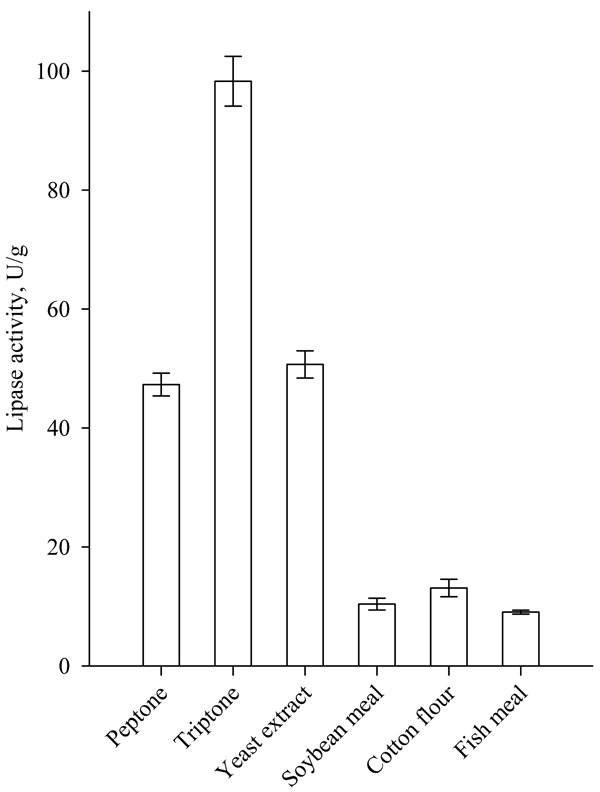
The highest lipase activity (about 270U/g) was achieved, when tryptone was added to the nutrient medium in concentration of 5.0% (w/w) (Fig. 6).

3.4. Effect of Particles Size and Moisture Content on Lipase Biosynthesis
The particles size of the solid substrate has a significant impact on the structure of the fermentation medium and hence on the possibility for mass transfer in the solid medium and on the growth of the strain. The effect of the size of wheat bran particles on lipase production is presented in Table 3.
| Particles Size | Lipase Activity, U/g |
|---|---|
| Reference | 288.53 |
| >1 mm | 249.88 |
| 0.2-1 mm | 191.75 |
| <0.2 mm | 56.72 |
With particle size reduction, mass transfer in the solid medium was most likely to be impaired, which significantly reduced lipase activity.
Another factor that significantly influences lipase biosynthesis is the moisture of the medium. Typically, the optimum moisture for growth of fungi strains in SSF is in the range of 50-70% w/w [18-20]. Highest lipase activity of 437.25 U/g was achieved at 60% moisture (Fig. 7). Moisture higher than 70% led to a decrease in lipase activity, due to a change in the structure of the medium resulting in porosity decrease, which hinders mass transfer and the strain development. Ferraz et al. have received similar results and determined that the optimal moisture for lipase production by Sporobolomyces ruberrimus was 60% [18].
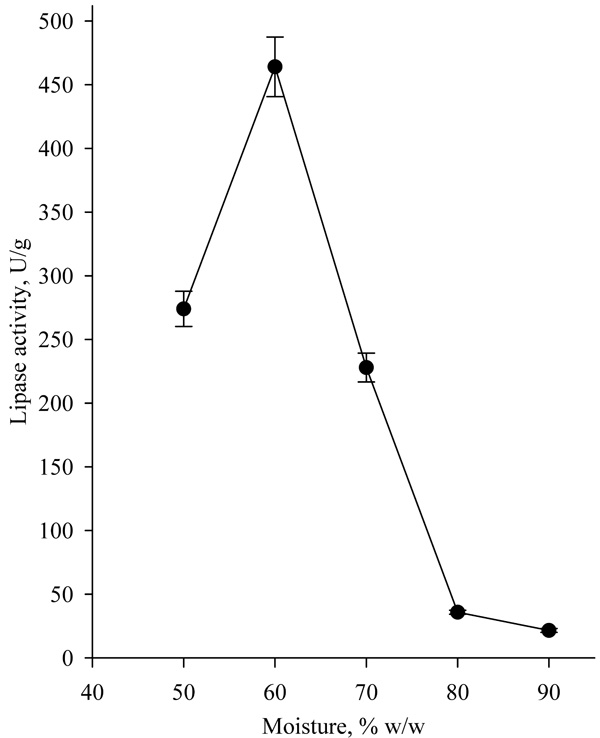
3.5. Optimization of Lipase Production by Response Surface Methodology
Of the studied factors, tryptone concentration and moisture content of the solid medium had the most significant effect on lipase biosynthesis. For these reasons, a mathematical experimental design was conducted for deveplopment of mathematical model to study the co-influence of both factors on lipase biosynthesis. The matrix of 22 OCD is shown in Table 4.
| Nº | Coded Levels | Lipase Activity, U/g | Predicted Lipase Activity, U/g | |
|---|---|---|---|---|
| Moisture (x 1 ) | Tryptone (x 2 ) | |||
| 1 | -1 | -1 | 74.68 | 74.84 |
| 2 | 0 | -1 | 400.00 | 392.27 |
| 3 | +1 | -1 | 167.74 | 175.31 |
| 4 | -1 | 0 | 135.96 | 141.57 |
| 5 | 0 | 0 | 437.25 | 440.84 |
| 6 | +1 | 0 | 214.90 | 205.71 |
| 7 | -1 | +1 | 110.00 | 104.24 |
| 8 | 0 | +1 | 381.20 | 385.34 |
| 9 | +1 | +1 | 130.41 | 132.04 |
The coefficients with a P-value <0.05 had a significant effect on lipase activity (Table 5). The mathematical model, describing the effect of moisture and tryptone concentration on lipase biosynthesis in SSF was developed as follows:
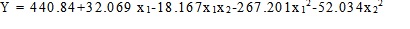 |
(2) |
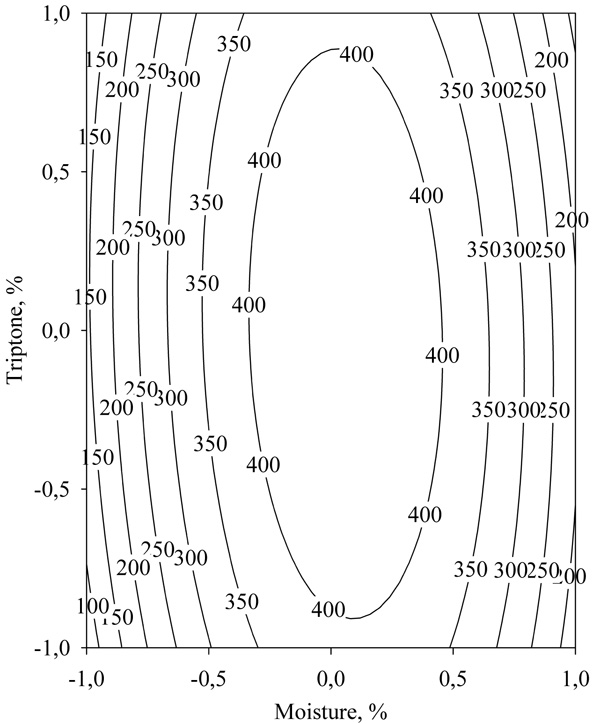
By studying the response surface (Fig. 8) it was found that the maximum lipase activity predicted by equation (2) was 441.81 U/g.
| Coeficient | Value | P-value |
|---|---|---|
| Intercept | 440.84 | 1.06E-05 |
| x1 | 32.06883 | 0.004273 |
| x2 | -3.46817 | 0.457192 |
| x1x2 | -18.1668 | 0.035725 |
| x12 | -267.201 | 4.05E-05 |
| x22 | -52.0335 | 0.005157 |
The optimal levels of the studied independent variables were 66% moisture (coded value 0.06) and 5.0% tryptone (coded value 0.01).
With the optimum values of moisture and tryptone concentration, 5 consecutive SSF experiments were performed, and the resulting lipase activity was 418.88 U/g ± 40.51. The achieved lipase activity was close to the value predicted by the model, which verified the adequacy of the model.
3.6. Lipase Extraction from the Fermented Solids
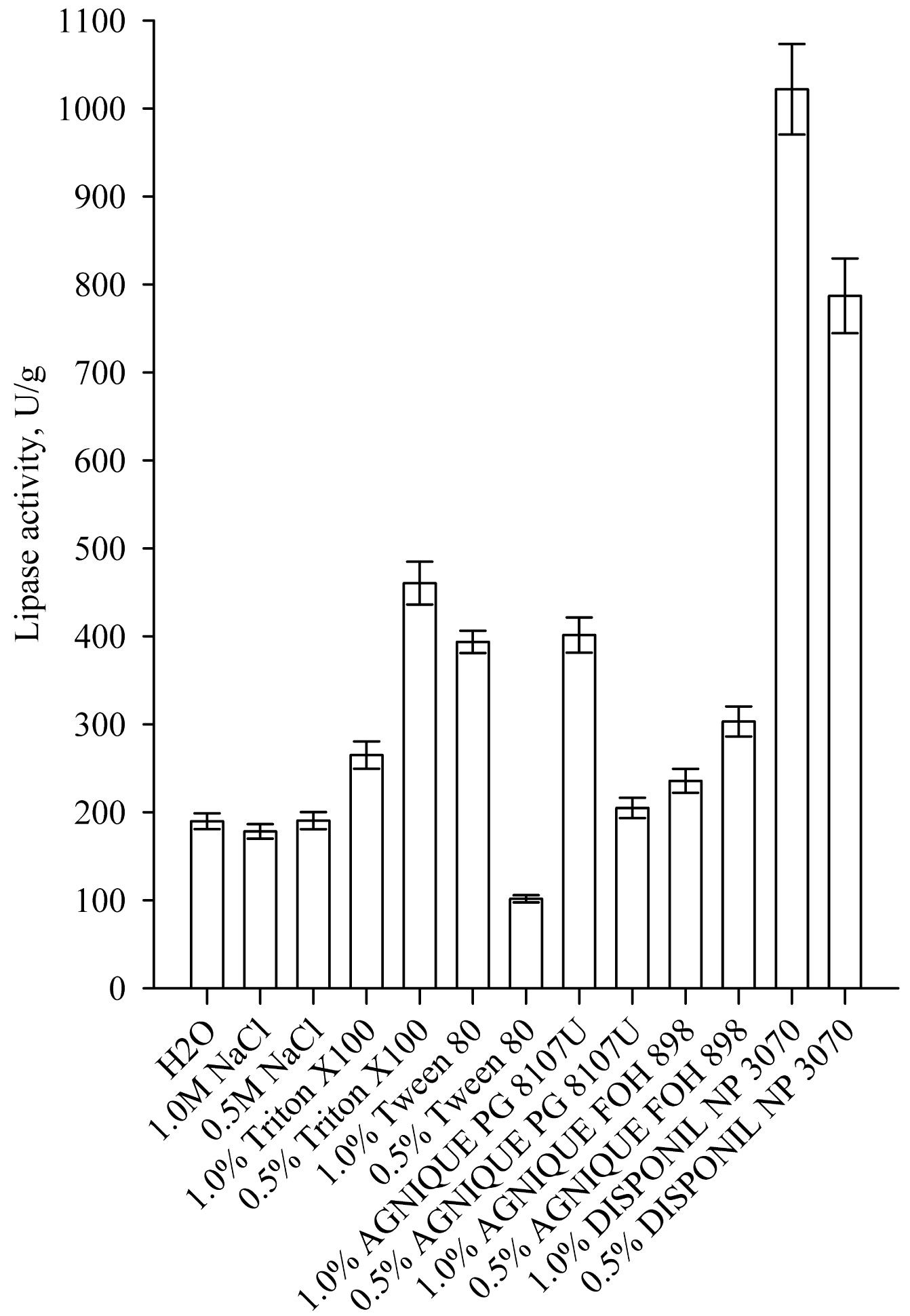
One of the processes that has a significant impact on enzyme yield in SSF is the enzyme extraction procedure from the fermented solids. Most often, to improve the extraction of lipase produced by SSF, nonionic surfactants are used, which do not denature the enzymes in contrast to most anionic and cationic surfactants. The results for the effect of different eluents for enzyme extraction on lipase activity is shown in Fig. (9). Highest lipase activity was achieved at 1.0% of Disponil NP 3070, which is a non-ionic coemulsifier. The lipase activity was 1021.80 U/g and it was almost 3 times higher than the activity of the reference sample with 1% Tween 80. Similar results were reported by other authors. Silva et al. found that the addition of nonionic surfactant Tween 80 increased lipase activity in the extract by 2.5 fold [4]. Rodrigez et al. found that the use of 1% Triton X-100 increased nearly 10 times the activity of lipase in the extract after SSF [21].
Lipase biosynthesis by Rhizopus arrhizus in SSF was compared with the process by other fungal strains grown on agricultural wastes (Table 6). The enzyme activity of lipase obtained after optimization of SSF conditions and extraction procedure (1021.80 U/g) was comparable and even higher than the values cited in the literature.
| Microorganism | Solid Substrate | Moisture,% | Conditions | Extraction | Lipase Activity, U/g | References |
|---|---|---|---|---|---|---|
| Rhizopus arrhizus | Wheat bran + tryptone | 66 | 168 h, 30oC |
1% Agnique | 1021.80 | In this work |
| Penicillium simplicissimum | Babassu cake + molasses |
65 | 72 h, 30oC |
0.1% Tween 80 | 85.7 | [4] |
| Rhizopus oligosporus | Almond meal + Tween 80 | 45 | 48 h, 30oC |
Phosphate buffer pH 7 | 81.22 | [6] |
| Aspergillus sp. | Soybean meal + rice husk | 60 | 12 days, 30oC |
0.2 M pH 7.0 Phosphate buffer |
25.07 U | [19] |
| Rhizopus homothallicus | Sugar-cane bagasse | 75 | 12 h, 40 oC |
20mM Tris–HCl buffer (pH 8), with 0.5% Triton X-100 and 2mM benzamidine |
1500 | [5] |
| Aspergillus niger | Wheat bran | 65 | 7 days, 30oC |
100 mL of distilled water | 9.14 | [7] |
| Penicillium simplicissimum | Babassu cake | 70 | 72 h, 30oC |
Phosphate buffer (100 mM, pH 7.0 | 19.6 | [20] |
| Penicillium sp | Soybean meal | 55ml/100g | 48 h, 27oC, |
0.1M Phosphate buffer, pH 7.0 | 200 | [22] |
| Rhizopus microsporus | Sugarcane bagasse+ wheat bran | 84 | 18 h, 40oC |
- | 262 | [15] |
| Rhizopus homothallicus | Sugarcane bagasse | 75 | 12 h, 40oC |
1% Triton-X-100 | 1224 | [21] |
| Aspergillus versicolor CJS-98 | Jatropha seed cake | 40 | 5 days, 25oC |
Chilled distilled water | 1079.47 | [23] |
CONCLUSION
The optimal conditions for production of lipase in SSF by Rhizopus arrhizus were determined. Maximum lipase activity was achieved when the strain was grown on wheat bran as solid substrate. Addition of glucose at a concentration of 1% (w/w) as an additional carbon source and tryptone at a concentration of 5% (w/w) as a source of organic nitrogen, significantly stimulated lipase biosynthesis. Using response surface methodology, the optimum moisture of the medium was determined to be 66% and the optimum tryptone concentration was 5% (w/w). As a result of the optimization, 418.88 U/g lipase activity was achieved. The procedure for enzyme extraction from the fermented solids had a significant effect on lipase yield. The addition of the non-ionic surfactant Disponyl NP 3070 in the eluent increased lipase activity about three folds. The achieved activity of 1021.80 U/g was higher and comparable with the activity of lipases, produced by other fungal strains. The optimization of the conditions and the use of low cost components in SSF makes the process economicaly effective for production of lipase from the investigated strain Rhizopus arrhizus.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies in this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The research was financially supported by Bulgarian National Science Fund (BNSF), Project 17/25-2017.


