All published articles of this journal are available on ScienceDirect.
Constitutive Expression of GATA4 Dramatically Increases the Cardiogenic Potential of D3 Mouse Embryonic Stem Cells
Abstract
The transcription factor GATA binding protein 4 (GATA4) is a vital regulator of cardiac programming that acts by inducing the expression of many different genes involved in cardiomyogenesis. Here we generated a D3 mouse embryonic stem cell line that constitutively expresses high levels of GATA4 and show that these cells have dramatically increased cardiogenic potential compared to an eGFP-expressing control cell line. Embryoid bodies (EB) derived from the D3-GATA4 line displayed increased levels of cardiac gene expression and showed more abundant cardiomyocyte differentiation than control eGFP EB. These cells and two additional lines expressing lower levels of GATA4 provide a platform to screen previously untested cardiac genes and gene combinations for their ability to further increase the efficiency of cardiomyocyte differentiation beyond that achieved by transgenic GATA4 alone. Non-integrative delivery of identified gene combinations will aid in the production of differentiated cells for the treatment of ischemic cardiomyopathy.
INTRODUCTION
The temporospatial expression pattern of transcription factors plays a vital role in cell lineage determination in the developing embryo. Commitment of a subset of embryonic cells to a cardiac fate is one of the earliest events in embryogenesis and involves GATA4, considered the master regulator of cardiogenic transcription [1]. GATA4 is expressed throughout cardiac development and adulthood [2], and loss of GATA4 function is incompatible with life [3].
GATA4 is a zinc finger transcription factor that binds to and activates the promoters of many cardiac-specific genes to direct the ordered proliferation, migration, and differentiation of cells in the precardiac mesoderm to form the embryonic heart. GATA4 has an N-terminal zinc finger that stabilizes protein-DNA interactions and a C-terminal zinc finger that mediates DNA binding at (a/t)GATA(a/g) residues and participates in interactions with co-stimulatory transcription factors such as SMADs, NKX2-5, and TBX5 [4-9]. The GATA4 sequence is highly conserved among mammals, underscoring its essential role in the development and function of the heart.
Cardiovascular disease is the leading cause of morbidity and mortality across the globe. The adult mammalian heart lacks significant regenerative capacity, and currently no therapies exist to replace damaged heart tissue. Cell-based treatment for cardiac ischemia offers great promise, but much needs to be learned before it can become clinically relevant. Pluripotent stem cells are theoretically ideal for cell-based therapies because they can differentiate into any cell type in an adult organism, enabling them to regenerate any tissue or organ. However, transplantation of undifferentiated pluripotent stem cells carries the potential for the formation of teratomas, rapidly proliferating tumors containing cell types from all three germ layers [10]. To avoid this concern, various protocols have been developed to differentiate stem cells into cardiomyocytes in vitro prior to transplantation [11-14]. The utility of these procedures for the generation of clinically applicable material ultimately hinges on their efficiency.
We are interested in using ectopic expression of essential cardiac transcription factors to augment cardiomyocyte differentiation from embryonic stem cells. Because GATA4 sits atop the hierarchy of the cardiac gene transcriptional network, it is logical to first record its effects in isolation prior to using it in combination with other factors. Accordingly, here we describe the creation of D3 mouse embryonic stem cell (mESC) lines that constitutively express human GATA4 cDNA. We characterized two of these lines and found that both displayed significantly greater cardiogenic potential than a control line despite widely varying levels of the human protein. Examination of the effects of constitutive GATA4 overexpression on endogenous cardiac gene activity revealed substantially increased levels of mouse GATA4 as well as NK2 homeobox 5 (Nkx2-5), cardiac troponin I (cTnI), and myosin light chain 2a (Mlc2a) transcripts in late-stage embryoid bodies (EB). These results suggest that our cell lines will be suitable for the exploration of additional cardiac developmental genes, potentially including GATA4-independent genes, that may complement GATA4 activity and thereby further enhance cardiomyogenesis.
MATERIALS AND METHODS
Cells and Culture Conditions
D3 mouse embryonic stem cells (ATCC CRL-11632) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning) supplemented with 15% fetal bovine serum (FBS, GIBCO), 1% non-essential amino acids (GIBCO), 1 mM sodium pyruvate (GIBCO), 100 U/ml penicillin (Cellgro), 100 µg/ml streptomycin (Cellgro), 0.1 mM 2-mercaptoethanol (GIBCO), and 1,000 U/ml mouse leukemia inhibitory factor (LIF, Millipore) (ES media) in 10 cm dishes coated with 0.2% gelatin (Sigma). Media was exchanged daily and the cells were passaged with trypsin every 2-3 d to maintain a state of self-renewal and minimize spontaneous differentiation.
Construction of GATA4 Expression Cassette
A GATA4 expression cassette consisting of the fusion cytomegalovirus (CMV) enhancer-chicken β-actin promoter (CAGp), the human GATA4 coding sequence, and the SV40 polyadenylation signal (SVpA) was constructed as follows. Plasmid pCDH-CMV-MCS-EF1Puro (System Biosciences) was modified by replacement of the resident CMV promoter between SpeI and EcoRI sites with a 1.7 kb CAG promoter SpeI-EcoRI fragment from plasmid pPEP100 (kindly provided by P. Spear, Northwestern University) [15] to create plasmid pCDH-CAG. A 1.4 kb EcoRI-SwaI fragment containing the human GATA4 cDNA was excised from pTopo-GATA4 (Open Biosystems) and inserted between the EcoRI and SwaI sites of pCDH-CAG to create pCAG-GATA4-EF1p-Puro. Plasmid pCAG-GFP-EF1p-Puro containing the eGFP coding sequence instead of the GATA4 sequence was created as a control.
Generation of D3 mESC lines expressing GATA4 and eGFP
Plasmids pCAG-GATA4-EF1p-Puro and pCAG-GFP-EF1p-Puro were digested at their unique SpeI site and the linearized material was gel-purified. D3 mESC were fed and 4 h later, the cells were detached with trypsin, pelleted, washed with PBS, and resuspended in 90 µl Ingenio Electroporation Solution (Mirus) per 3x106 cells. 10 µg linearized DNA diluted with 10 µl Ingenio Electroporation Solution was mixed with the cells in an electroporation cuvette and the DNA was nucleofected into the cells using an Amaxa Nucleofector I set to program A-13 (bio.lonza.com/ fileadmin/groups/marketing/Downloads/Protocols/Generated/Optimized_Protocol-103.pdf). Immediately after nucleo- fection, the cells were diluted by the addition of 500 µl warm media and plated in gelatin-coated 10 cm dishes containing warm ES media with LIF. Media was changed 24 h after plating and on the following day, 2 µg/ml puromycin was added to the media. Puromycin-resistant colonies began to appear 24 h after the onset of selection and were picked under a microscope in a sterile environment one week after nucleofection. Each colony was placed in one well of a 24-well plate for expansion. A minimum of 10 colonies were expanded from each nucleofection. Selected clones referred to as D3-eGFP (eGFP nucleofection) or D3G4 (GATA4 nucleofection) were characterized by observation of eGFP expression (D3-eGFP clones), Western blotting (D3G4 clones), OCT4 indirect immunofluorescence with mouse monoclonal antibody sc-5279 (Santa Cruz; 1:100 dilution) and Alexa Fluor 488 donkey anti-mouse IgG (ab150105, Abcam; 1:1,000 dilution), EB assay, and quantitative real time polymerase chain reaction on reverse transcribed mRNA (qRT-PCR).
Western Bot Analysis
Cell lysates were collected from undifferentiated D3G4 mESC lines in RIPA lysis buffer supplemented with protease inhibitors (Roche). Equal amounts of protein were loaded on a 10% polyacrylamide gel and after electrophoresis, were transferred by semi-dry transfer onto a PVDF membrane (Millipore). The membrane was washed in Tris-buffered saline (TBS) (20 mM Tris, 0.5 M NaCl, pH 7.5), blocked for 1 h at room temperature in TBS-T (0.5% [vol/vol] Tween 20 in TBS) supplemented with 10% (vol/vol) nonfat dry milk, and incubated overnight at 4°C with a 1:100 dilution of anti-GATA4 antibody ab84593 (Abcam) in blocking buffer. The following day, the membrane was washed 3 times for 10 min each in TBS-T, incubated with a 1:10,000 dilution of horseradish peroxidase-conjugated anti-mouse secondary antibody (RABHRP1, Sigma) diluted in blocking buffer for 1 h at room temperature, and developed using SuperSignal™ West Dura Chemiluminescent Substrate (Thermo Scientific).
Embryoid Body Assay
Cells were suspended in ES media without LIF at a concentration of 4x104 cells/ml. 20 µl drops of cells were pipetted onto the inverted lid of a petri dish, and the lid with the attached drops was turned back over and placed on the dish containing 30 ml sterile water to keep the drops from evaporating. The cells were incubated at 37°C in an atmosphere of 5% CO2 to promote aggregation into EB and initiate differentiation. After 48 h in hanging drops, the EB were pipetted one per well into 48-well plates, each well containing 500 µl ES media without LIF. Starting on day 8 (d8) from the start of EB formation, the number of wells containing spontaneously contracting cells was recorded daily for up to 7 days.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from undifferentiated D3 cells, 12d-old EB formed by D3-eGFP and D3G4 line B cells, and adult mouse heart using the Qiagen RNeasy Plus mini kit with gDNA eliminator columns (Qiagen). Reverse transcription was performed with the Ambion RETROscript Kit (Ambion) on 1-2 µg RNA using random decamers as primers (Ambion) in a final volume of 20 µl. The resulting cDNA was diluted 1:10 for qPCR analysis.
Primers for qPCR (Invitrogen) are listed in Table 1. qPCR was performed using LightCycler SYBRgreen Master Mix (Roche) and a Step One Plus Real-Time PCR System (Applied Biosystems). Cycling conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. All qPCR reactions were performed in triplicate and the results for each cDNA were averaged. Threshold cycle (Ct) values were used to calculate changes in gene expression using the 2-ΔΔCt method [16]. Results were normalized to Hprt cDNA and fold change was calculated relative to undifferentiated D3 cells. Final qRT-PCR products were visualized on a 2% agarose gel by ethidium bromide staining.
qRT-PCR primers.
| Gene | Forward Primer | Reverse Primer | Product Length (bp) |
Reference |
|---|---|---|---|---|
| GATA4 (human) | TTCCAGCAACTCCAGCAACG | GCTGCTGTGCCCGTAGTGAG | 97 | [17] |
| GATA4 (mouse) | GCCCAAGAACCTGAATAAAT | CGGACACAGTACTGAATGTCT | 195 | [18] |
| Nkx2-5 | CAGTGGAGCTGGACAAAGCC | TAGCGACGGTTCTGGAACCA | 217 | [19] |
| Mlc2a | TCAGCTGCATTGACCAGAAC | AAGACGGTGAAGTTGATGGG | 148 | [20] |
| cTnI | ACGTGGAAGCAAAAGTCACC | CCTTCTTCACCTGCTTGAGG | 189 | [18] |
| cTnT | GAGGAACAGGAAGAGCAAGA | ACCAAGTTGGGCATGAAGAG | 191 | [18] |
| Hprt | GCTGGTGAAAAGGACCTCT | CACAGGACTAGAACACCTGC | 249 | [21] |
| Oct4 | TGTTCCCGTCACTGCTCTGG | TTGCCTTGGCTCACAGCATC | 82 | [22] |
RESULTS
Generation of GATA4 and Control mES Cell Lines
We constructed dual gene expression plasmids containing human GATA4 cDNA or the eGFP gene under transcriptional control of the fusion cytomegalovirus (CMV) enhancer-chicken β-actin promoter (CAGp) and the puromycin resistance gene under control of the EF1α promoter (Fig. 1A). Following linearization, the plasmids were introduced into D3 mESC by nucleofection and transduced colonies were identified by selection for resistance to puromycin. Several individual colonies of each kind were isolated and expanded.
Clonal lines expressing eGFP (D3-eGFP) retained normal stem cell morphology and every colony formed upon low-density seeding of the cells expressed eGFP (Fig. 1B). We performed a preliminary screen of 13 lines from the GATA4 plasmid nucleofection. Upon differentiation (see next section), all lines showed increased numbers of beating centers. Ten were screened by Western blot for GATA4 expression, showing a range of signals from undetectable to strong. We chose 3 lines representing low, intermediate and high expression, referred to as D3G4-C, -A, and -B, respectively, and confirmed their expression levels by repeat Western blot. As shown in Fig. (1C), D3G4 line B expressed abundant GATA4, line C appeared negative, and line A showed an intermediate level of the protein. No GATA4 was detected in unmodified D3 mESC or the D3-eGFP line (data not shown).
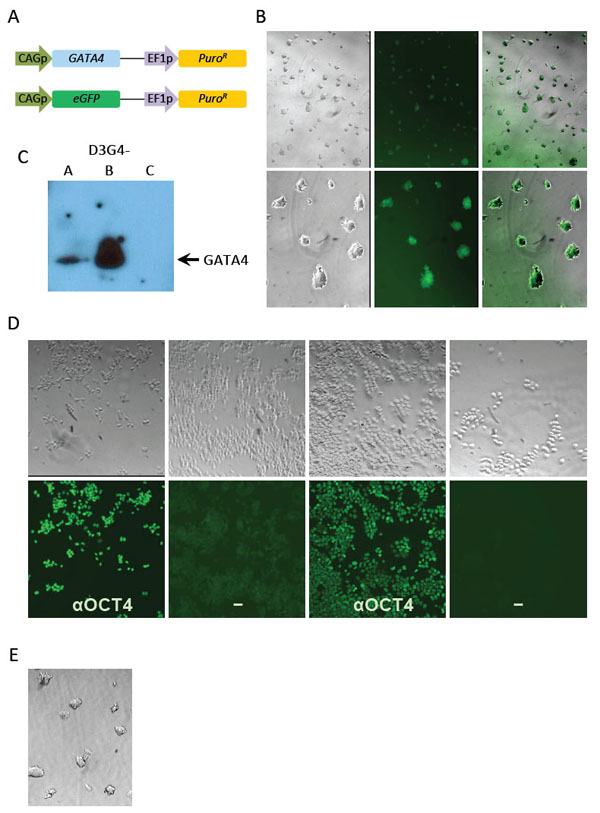
Generation of GATA4- and eGFP-expressing D3 mESC cell lines. (A) Dual expression cassettes in nucleofected GATA4 (top) and eGFP (bottom) plasmids. CAGp, CAG promoter; EF1p, EF1α promoter; PuroR, puromycin-resistance gene. (B) eGFP expression in undifferentiated cells from a D3-eGFP clone at low (upper panels) and high (lower panels) magnification. Cells are shown from left to right in brightfield, fluorescence, and overlay. (C) Western blot for GATA4 detection in three D3G4 clonal lines. (D) Cloned D3-eGFP cells (4 panels at left) and D3G4 clone B cells (4 panels at right). Upper row, brightfield; lower row, the corresponding fields stained with anti-OCT4 and fluorescent secondary antibody (αOCT4) or secondary antibody alone (-). (E) Brightfield image of undifferentiated D3G4-B cells at high magnification as in (B).
We used indirect immunofluorescence to evaluate the persistence of pluripotency marker OCT4 in the highest GATA4-expressing clone, D3G4-B, and control D3-eGFP cells after several passages in LIF-containing media. Essentially all cells stained positive in the presence of OCT4 antibody (Fig. 1D, αOCT4 panels) while no signal was apparent in D3G4-B cells in the absence of primary antibody (Fig. 1D, lower right); D3-eGFP cells showed faint green fluorescence in the absence of primary antibody due to eGFP expression (Fig. 1D, second from left). Consistent with these results, D3G4-B (Fig. 1E) as well as clones A, C and D3-eGFP appeared morphologically stable through multiple passages in LIF-containing media although increased spontaneous differentiation was observed for all clones at late passages (>10, data not shown). Accordingly, we used only low-passage cells in the remainder of this study.
Ectopic GATA4 Enhances Cardiomyocyte Generation
To determine whether constitutive expression of human GATA4 affects mESC differentiation, we generated EB from unmodified D3 cells, D3-eGFP, and D3G4 lines B and C by cell aggregation in hanging drops. After 48 h, we transferred each aggregate into a separate well of a 48-well culture plate and from days 8-14 after the start of cell aggregration (onset of differentiation), we monitored the EB (n=48/cell line) daily for the appearance of spontaneously contracting cells representing mature cardiomyocytes. While few D3 and D3-eGFP EB exhibited evidence of cardiomyocyte formation, many EB derived from the two D3G4 lines displayed readily detectable areas of contracting cells (Fig. 2). D3G4 line B, expressing the highest GATA4 level, had the largest percentage of EB with contracting cells (90% on day 12). Surprisingly, while line C appeared to express little or no GATA4 protein, almost 65% of EB from this line showed areas of contracting cells at day 14 compared to a maximum of 17% of control EB at any time during the observation period.
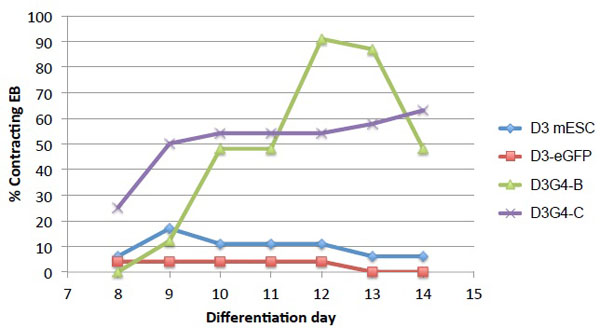
Cardiomyocyte differentiation of transduced mESC lines. Control D3 cells, line D3-eGFP, and D3G4 lines B and C were differentiated in EB (48/cell line). EB with visible areas of contracting cells were counted daily on days 8-14 post initial cell aggregation.
GATA4 Transduction Stimulates the Expression of Cardiac-Specific Genes During Differentiation
We used qRT-PCR to examine the expression of selected cardiac genes in late-stage EB derived from D3-eGFP and D3G4 line B compared to undifferentiated D3 cells and adult mouse heart. Since the previous experiment (Fig. 2) showed the highest percentage of beating D3G4 EB on day 12, we chose this day for RNA extraction from pools of EB. In this instance, 74% of EB from line B contained contracting foci on day 12 while none of the D3-eGFP EB did (Fig. 3A). As expected, expression of the pluripotency marker gene Oct4 was dramatically downregulated in d12 EB compared to undifferentiated D3 cells and was undetectable in mouse heart cDNA (Fig. 3B). We used species-specific primers to distinguish human GATA4 and mouse GATA4 cDNAs. Although the human-specific primers showed background amplification from both D3-eGFP EB and mouse heart cDNA, the signal from D3G4 line B cDNA was more than 100-fold higher. Expression of the mouse GATA4 gene, as well as the Nkx2-5 and cTnI genes, was up to ~10-fold higher in EB from line B than in D3-eGFP EB, and Mlc2a gene expression was over 100-fold increased in line B EB compared to EB from D3-eGFP cells. Previous studies have shown that GATA4 and Nkx2-5 are expressed throughout cardiomyogenesis while cTnI and Mlc2a are expressed only at the later stages [23-25]. Visualization of the qRT-PCR products of these and another cardiac differentiation gene, cTnT, indicated that the expression of these genes in D3G4 EB more closely resembled that in mouse heart than in D3-eGFP EB (Fig. 3C).
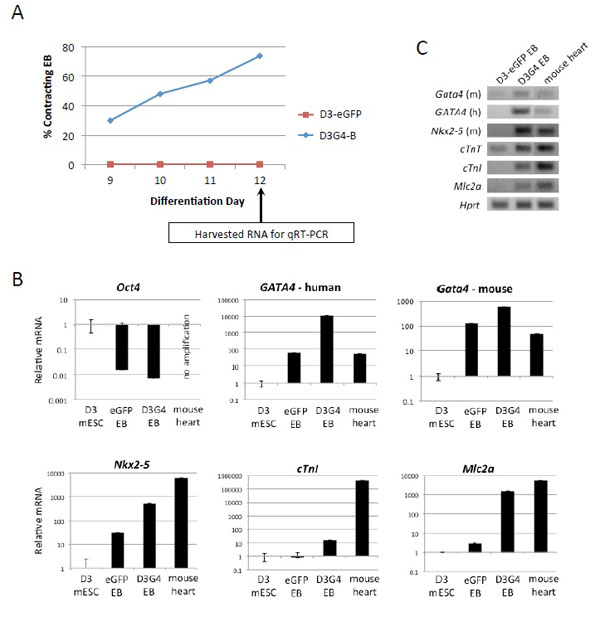
Cardiac developmental gene expression in EB from D3-eGFP and D3G4 line B. (A) Percentage of EB (n=19/cell line) containing microscopically detectable areas of contracting cells on days 9-12 past the onset of differentiation. The time of RNA extraction for (q)RT-PCR analysis is indicated. (B) mRNA levels for the indicated genes as measured by qRT-PCR. Data was normalized to mouse Hprt cDNA and is expressed relative to undifferentiated D3 mESC. (C) qRT-PCR products identified by agarose gel electroporesis and ethidium bromide staining (m, mouse; h, human).
DISCUSSION
The goal of this study was to create mES cell lines that constitutively express GATA4 and can be used to screen other ectopic genes for their ability to stimulate cardiomyocyte generation beyond the level achieved by GATA4 alone. Previous reports have shown that GATA4 acts cooperatively with other early cardiac transcription factors to activate cardiac developmental gene expression [5, 7, 8], and ectopic expression of combinations of GATA4 with some of these factors has been shown to increase mESC differentiation into cardiomyocytes [26-28]. However, less attention has been paid to factors acting at later stages whose expression may not be fully responsive to these early regulators. While GATA4 is considered a master regulator of cardiogenesis, clearly it does not act alone and its overexpression may therefore cause imbalances throughout the developmental gene expression program. Thus we wished to establish a system that can be used to rapidly test different types of cardiac genes for their ability to enhance GATA4-induced cardiomyogenesis, potentially by correcting imbalances in the gene expression program. To this end, we created D3 mouse embryonic stem cell lines that constitutively express different levels of human GATA4. Using an eGFP-expressing line as a control, we characterized one of the lines in some detail, showing its enhanced cardiogenic potential. We recorded the appearance of spontaneously contracting areas in EB cultured for two weeks following the onset of differentiation and examined the expression of several cardiac-specific genes in 12 day old EB. In addition, we identified 2 other lines, expressing lower or undetectable levels of GATA4, that will allow us to explore GATA4 dosage effects on combinatorial transduction approaches.
Although all transduced D3 cell lines were maintained under puromycin selection, GATA4 protein levels in three D3G4 lines varied widely. No GATA4 was detected in line C, while line B produced robust amounts of the protein and line A showed an intermediate level. Visual inspection of differentiating EB showed that 75-90% of EB from line B developed large areas of spontaneously contracting cells while fewer than 20% of EB derived from unmodified D3 or D3-eGFP cells contained any areas of contracting cells. Despite the absence of detectable GATA4 protein in line C, many more EB from this line displayed contracting areas (maximum 63%) than from the controls. This may indicate that a small amount of ectopic GATA4 that is below the limit of detection by Western blot is sufficient to enhance cardiomyocyte differentiation from mESC. Alternatively, line C may have acquired other changes during its isolation and propagation that predisposed the cells to cardiomyogenesis, perhaps explaining the earlier appearance of contracting EB from this line than from line B (Fig. 2).
Analysis of RNA from late-stage EB from D3G4 line B by quantitative RT-PCR compared to RNA from D3-eGFP EB showed increased expression of endogenous GATA4 and Nkx2-5, key initiators of cardiac differentiation that are expressed throughout cardiomyogenesis [3, 23], and several genes that are mainly expressed in mature cardiomyocytes [24, 25]. It is known that GATA4 upregulates its own expression as well as that of NKX2-5 [5, 29-31], and these two proteins interact to activate the promoters of other cardiac differentiation genes [7, 8-32]. Our results indicate that ectopic GATA4 expression can enhance this gene expression cascade, resulting in increased yields of mature cardiomyocytes from mESC without extensive manipulation of culture conditions, such as the timed addition of stimulatory factors and intermittent cell sorting. It is likely, however, that such manipulations as well as co-expression of other cardiac transgenes can further enhance cardiomyocyte production.
Cell lines generated by stable transfection can show wide variations not only in transgene expression levels, but also in other characteristics. For example, we found that several lines, including D3G4 line A, were much more prone to spontaneous differentiation than others. This may be due to differences in transgene integration sites that can differentially affect both transgene and endogenous gene expression, potentially resulting in selection of different secondary events during propagation of the cells. In turn, these events may cause variations in growth rates and spontaneous differentiation between individual lines. Thus, generalizations based on observations from a few cell lines must be viewed with caution. Nonetheless, our findings confirm that forced GATA4 expression provides a means to stimulate the cardiogenic potential of mESC.
Previous studies have described GATA4 as the master regulator of cardiogenesis [3, 23] and have shown GATA4 expression in vivo throughout all developmental stages from precardiac mesoderm to the adult heart [2]. The evidence presented here and elsewhere [33, 34] that persistent overexpression of GATA4 can drive pluripotent stem cells toward a cardiac lineage without detrimental effects on terminally differentiated cardiomyocytes is consistent with these prior observations. However, the approach taken in our study is not suitable for the generation of clinically applicable cardiomyocytes because stable transfection causes permanent chromosomal alterations, raising the risk of oncogenesis [35]. Instead, nonintegrative methods of gene delivery will be required to use forced GATA4 and supplemental transgene expression for the generation of a clinically relevant product. Our observation that human GATA4 induces expression of its mouse counterpart as well as other cardiac genes, directly or indirectly, raises the possibility that an early pulse of ectopic GATA4 expression may be sufficient to commit daughter cells to the cardiac lineage. Thus it will be worthwhile to determine whether transient ectopic GATA4 expression from a replication-defective, non-integrating viral vector can enhance cardiomyocyte production from pluripotent stem cells. The cell lines described here will help us identify genes that may aid in securing the proper balance of gene expression through the remaining developmental stages.
CONCLUSION
Stable expression of the human version of the early cardiac transcription factor GATA4 in D3 mouse embryonic stem cells resulted in increased cardiomyocyte formation in differentiating embryoid bodies and increased expression of endogenous cardiac differentiation genes. These cell lines will be valuable to identify previously untested genes for their ability to act cooperatively with GATA4 to enhance cardiomyogenesis. The long-term goal of this work is to deliver differentiation-enhancing gene combinations extra-chromosomally for cell therapy of ischemic cardiomyopathy.
ABBREVIATIONS
| CAGp | = CMV enhancer-chicken β-actin promoter |
| CMV | = Cytomegalovirus |
| cTnI | = Cardiac troponin I |
| cTnT | = Cardiac troponin T |
| EB | = Embryoid bodies |
| EF1α | = Elongation factor 1α |
| GATA4 | = GATA binding protein 4 |
| Hprt | = Hypoxanthine-guanine phosphoribosyltransferase |
| LIF | = Leukemia inhibitory factor |
| mESC | = Mouse embryonic stem cell(s) |
| Mlc2a | = Myosin light chain 2a |
| Nkx2-5 | = NK2 homeobox 5 |
| qRT-PCR | = Quantitative reverse transcription PCR |
| SVpA | = SV40 polyadenylation signal |
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by NIH grants HL066949 and DK044935 to J.C.G. L.L.L. was supported in part by an NRSA Institutional Predoctoral Training Grant in Molecular Microbial Persistence and Pathogenesis.


