All published articles of this journal are available on ScienceDirect.
A Fusarium Oxysporum-derived Compound Exhibits Potential Insecticidal Activity against Cotton Bollworm: Evidence from In Silico and In Vitro Analyses
Abstract
Introduction
Fungal derived secondary metabolites serve as promising alternatives to traditional insecticides due to their eco-friendly nature and specific modes of action. This study investigates the insecticidal potential of a secondary metabolite derived from Fusarium oxysporum using in silico and in vitro approaches. The objectives include the purification of the bioactive compound, followed by computational and experimental evaluations to assess its insecticidal efficacy.
Methods
The initial purification involved mass spectroscopic analysis, followed by structural elucidation using Varian 500 NMR. The compound was tested for insecticidal potential using third instar nymphs of Helicoverpa armigera. From stock solution prepared in sterile dimethyl sulfoxide (DMSO) test concentrations of 100, 250, 500, and 1000 µg mL-1 were prepared. Imidacloprid in powder form was used as the standard insecticide in this experiment, also at concentrations of 100, 250, 500, and 1000 µg mL-1. Computational analyses, including molecular docking, molecular dynamics simulations and binding free energy calculations, were performed to explore the compound’s interaction with the target receptor.
Results
The compound derived from F. oxysporum was identified as 6-butyl-3-(1-hydroxyethyl)-2-methyl-2H-pyran-5-carbonyl carbamic acid having an exact mass of 283 and molecular formula C14H21NO5. The fungal compound, administered at concentrations of 100, 200, and 250 µg mL-1, exhibited mortality rates of 30%, 56.6%, and 63.3%, respectively. In comparison, the standard drug, imidacloprid, demonstrated mortality rates of 30%, 50%, and 60% at the respective concentrations. At a concentration of 500 µg mL-1, the fungal compound inhibited the growth of 8 larvae in each test plate, resulting in an 80% mortality rate while, the standard at this concentration caused the death of 8 larvae in two petriplates and 7 larvae in the third, resulting in a mortality rate of 76.6%. At the highest applied concentration (1000 µg mL-1), both the fungal compound and the standard exhibited 100% mortality by inhibiting the growth of every larva in all petriplates. The docking analysis predicted -5.3 binding energy that represents the selected compound having significant binding affinity for the target receptor. Similarly, the compound showed stable dynamics with the enzyme and reported robust binding energies.
Discussion
The findings highlight the potential of 6-butyl-3-(1-hydroxyethyl)-2-methyl-2H-pyran-5-carbonyl carbamic acid as a bioactive fungal metabolite for eco-friendly pest control, warranting further investigations into its mechanism of action, field efficacy, and environmental safety. Future work will focus on chemical synthesis to produce the compound in sufficient quantities for comprehensive biological evaluations, including detailed toxicity profiling, selectivity assessments and environmental safety studies. Additionally, field trials will be conducted to evaluate its real-world efficacy in pest management.
Conclusion
This study demonstrates that 6-butyl-3-(1-hydroxyethyl)-2-methyl-2H-pyran-5-carbonyl carbamic acid exhibits potent insecticidal activity against Helicoverpa armigera, with efficacy comparable to imidacloprid.
1. INTRODUCTION
Insect pest management remains a significant challenge in global agriculture, leading to widespread reliance on synthetic chemical pesticides [1, 2]. However, growing concerns regarding their environmental impact and risks to human health have prompted the search for safer and more sustainable alternatives [3]. The harmful effects of synthetic pesticides, extensively documented in the United Nations Environmental Program’s 1997 report, include threats to human and animal health and large-scale environmental contamination [4]. These concerns highlight the urgent need for eco-friendly pest control strategies that minimize chemical residues and reduce ecological imbalances [5, 6]. Fungal-derived metabolites have emerged as promising alternatives for insect control due to their biodegradability, target specificity, and reduced environmental toxicity [7]. These metabolites, produced as part of fungal secondary metabolism, exhibit precise modes of action with minimal collateral damage to non-target organisms [8]. Their ecological benefits, including potential synergistic interactions with other biological control agents, make them as valuable tools for sustainable pest management [9, 10].
The transition toward fungal-based insecticides requires a deeper understanding of their biochemical mechanisms and ecological effects. Advances in biotechnology and fermentation techniques have facilitated large-scale production and extraction of fungal metabolites, significantly enhancing their accessibility for agricultural applications [11]. Several fungal species, including Beauveria bassiana, Metarhizium anisopliae, and Paecilomyces spp., have demonstrated strong insecticidal properties against a diverse range of pests [12-14].
The integration of modern computational tools has revolutionized our understanding and application of fungal secondary metabolites [15, 16]. Techniques such as molecular modeling, simulation, and pharmacophore modeling enable researchers to predict the biological activities, molecular targets, and structural properties of fungal metabolites with exceptional precision [17, 18]. This multidisciplinary approach not only broadens our knowledge base but also paves the way for innovative and sustainable pest management strategies. Recent studies have highlighted the potential of fungal metabolites to not only act as potent insecticides but also contribute to the development of resistance management strategies by targeting specific biochemical pathways in pests [19]. Moreover, the ecological benefits of using fungal-derived insecticides are significant, as they often exhibit specificity towards target pests while being less harmful to non-target organisms and the environment. This aligns with the growing demand for sustainable agricultural practices that are environmentally friendly and reduce harm to nature [20]. The continuous exploration and characterization of fungal secondary metabolites, supported by advancements in computational biology and biotechnology, are essential for the next generation of eco-friendly and effective insecticides [21].
In the current study, we report the purification and characterization of a secondary metabolite derived from F. oxysporum with potential insecticidal properties. After purification and structural elucidation, the compound was subjected to molecular docking and simulation studies to predict its biological activity and molecular targets. Based on the in silico findings, the insecticidal potential of the compound was further evaluated through in vitro bioassays against Helicoverpa armigera.
2. METHODOLOGY
The flow diagram of the applied methodology is shown in Fig. (1).
2.1. Rhizosphere Sample Processing and Isolation of Fungal Strains
Samples were obtained from the soil surrounding the roots of ginger plants (Zingiber officinale) cultivated in the botanical garden of the Department of Botany, University of Peshawar. Fungal isolation was achieved through the soil dilution plate method, followed by inoculation of the samples onto Potato Dextrose Agar (PDA) for ten (10) days at 28°C [22]. Upon completion of the incubation period and visual inspection of fungal growth, the fungal strains were subsequently sub-cultured and purified.
2.2. Identification of the Bioactive Fungal Strain
The bioactive fungal strain was identified through a series of bioactivity tests, leading to its selection for further analysis and purification of secondary metabolites. Identification of fungal strains involved analyzing various traits, including hyphal structure, colony morphology, and the arrangement of conidia. Colony pigmentation was evaluated by cultivating the strains on different media, such as Potato Dextrose Agar (PDA), Czapec Dox Agar (CDA), and Saboraud Dextrose Agar (SDA). Hyphal morphology, spore structure, and arrangement were examined using a light microscope and the slide culture technique. To enhance visualization, slides were stained with lactophenol blue [23-25].
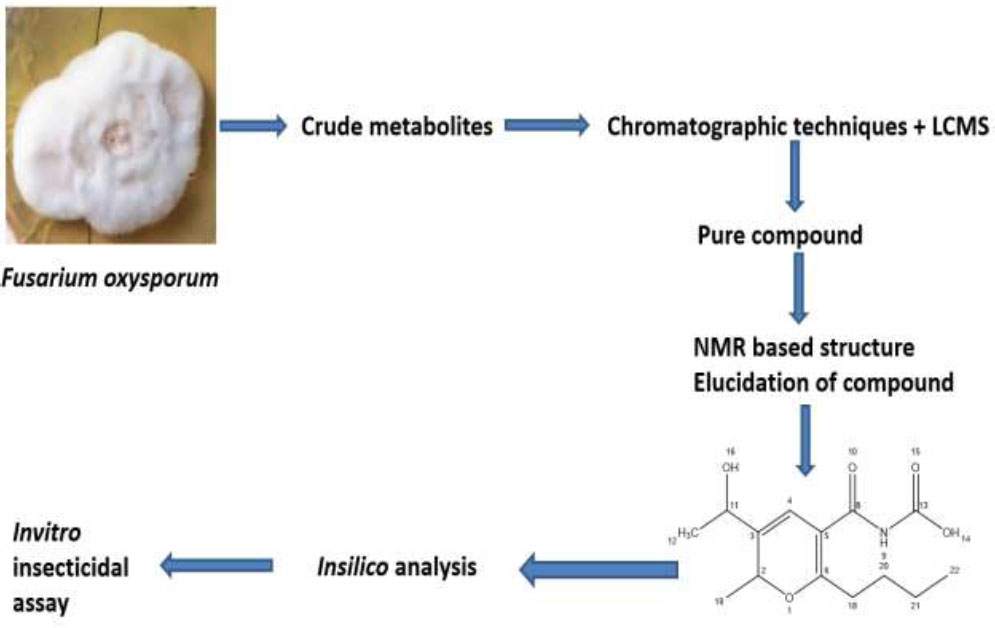
Schematic flow of the current study.
2.3. Fungal Metabolites Production and Extraction using Yeast Extract Supplemented (YES) Medium
For the production of crude metabolites, a fungal culture was cultivated in a modified yeast extract-supplemented (YES) medium. To formulate a liter of YES medium optimized for fungal metabolite production, yeast extract, peptone, and glucose were dissolved in distilled water at concentrations of 2% (w/v) each. Additionally, sucrose (2% w/v) was added to serve as an alternative carbon source, while ammonium sulfate (0.5% w/v) was introduced to augment nitrogen availability. Ensuring robust growth and metabolic activity, a trace element solution including iron sulfate (0.01% w/v), zinc sulfate (0.001% w/v), copper sulfate (0.0001% w/v), and manganese sulfate (0.0001% w/v) was incorporated. The pH of the medium was precisely adjusted to 5.8 before autoclaving at 121°C [26, 27]. Sterility confirmation was conducted through overnight incubation at 28°C. After inoculation with the desired fungal strain, the medium was transferred to a shaking incubator and maintained at 28°C and 150 rpm for 14 days.
To expedite the separation and sedimentation of media components, approximately 300 μL of 35% hydrochloric acid (HCl) was added to each flask. The mycelia were homogenized using an electric blender, after which an equal volume of ethyl acetate was introduced to each flask. The mixture was intermittently mixed and shaken for 30 minutes. The mycelia were then filtered using cheesecloth. The organic layer was separated from the resulting mixture using a separating funnel and washed with a 2M brine solution to remove impurities. Anhydrous sodium sulfate (Na2SO4) was employed to dehydrate the organic layer and remove any residual moisture [28, 29]. The organic layer, containing the metabolites in crude form, was filtered again with the help of filter paper and transferred to a rotary evaporator set at 45°C to concentrate the metabolites. The dried crude extract was collected from the rotary evaporator flask by rinsing it with methanol.
2.4. Purification and Analysis of Fungal Metabolites
For the purification and analysis of the fungal metabolites, a combination of equipment was employed. This included the Waters 2795HT HPLC system, linked with a Waters 2998 photodiode array detector and a Micro-Mass ZQ spectrometer, enabling liquid chromatography-mass spectrometry (LCMS) analysis. The UV detection range of the instrument spanned from 200 to 400 nm. The solvent system utilized for both analytical and preparative runs comprised HPLC-grade ultrapure water, HPLC-grade methanol (MeOH), and HPLC-grade acetonitrile (CH3CN), supplemented with a minimal quantity of formic acid (0.055%). Initially, a 20 µl injection was administered for the analytical run, with subsequent increases in volume for further purification processes.
2.5. NMR-based Structural Elucidation of the Fungal Pure Compound
One-dimensional and two-dimensional NMR spectra (1H, 13C, and HSQC) were acquired utilizing a 500 MHz Varian NMR machine. Chemical shifts, denoted in ppm, were calibrated using tetramethylsilane (TMS) as the internal standard. For examination, a solution comprising 5 mg of the pure compound dissolved in 0.65 mL of deuterated chloroform was prepared, and a series of one-dimensional and two-dimensional NMR experiments were carried out on a 500 MHz NMR spectrometer.
2.6. Molecular Docking Investigation
The molecular docking technique was then used to investigate the binding conformation and affinity of the compound for any given insecticidal macromolecule [30]. In this study, Glutathione S-transferase (PDB ID: 1R5A) from Anopheles cracens was used as a receptor [31]. The compound structure was drawn using ChemDraw 12.0 (Milne, G.W., 2010) and subsequently energy-minimized using UCSF Chimera 1.17 [32]. The enzyme receptor was also energy minimized using the steepest descent and conjugate gradient algorithms over a total of 3000 rounds. Both the compound and the enzyme receptor were saved as pdb files. Docking analysis was accomplished using PyRx 0.8 software [33]. The grid box around the enzyme active site was set considering dimensions of 30 Å along XYZ. For the compound, 100 poses were generated, and the top binding mode was selected based on negative binding energy. The complex was visualized using Discovery Studio v2021 [34].
2.7. Molecular Dynamics Simulation and Binding Free Energies Analysis
The dynamic movements of the docked complex were studied using molecular dynamics simulation through AMBER v22 software [35]. The complex initial processing was done using an antechamber program. The enzyme was treated using the FF19SB force field, while the compound was processed with GAFF2 [36, 37]. The simulation protocol was achieved using four phases: energy minimization, heating, equilibrium, and production run [38]. The temperature control during the production run was done using the Langevin algorithm, while the SHAKE was employed to constrain hydrogen bonds [39]. The CPPTRAJ module was used for plotting different simulation-based graphs [40]. The binding free energies were calculated using the MMPBSA.py module for over 10,000 frames from simulation trajectories [41].
2.7.1. In vitro Insecticidal Potential of the Pure Compound
Third-instar nymphs of H. armigera were obtained from the entomology department at the University of Agriculture Peshawar and placed in sterile jars containing synthetic medium. The larvae were reared in a controlled laboratory environment at 30°C and 75% relative humidity and fed a synthetic diet primarily composed of chickpea flour. The diet consisted of specific quantities of various components: 50 g of chickpea flour, 5 g of wheat germ, 12 g of yeast extract, 3.5 g of casein, 0.5 g of sorbic acid, and 1 g of methylparaben in 150 mL of distilled water for the mixture (A); 0.35 g of choline chloride, 0.02 g of streptomycin sulfate, 2 g of ascorbic acid, 0.15 g of cholesterol, a multivitamin multi-mineral capsule (manufactured by GlaxoSmithKline Pharmaceuticals Limited), 200 mg of vitamin E, 1 mL of formaldehyde, 0.3 g of bavistin, and 30 mL of distilled water for the mixture (B); and 6.5 g of agar in 180 mL of distilled water for mixture (C). Components A and B were combined and then mixed with molten agar (C). This medium was also used to conduct insecticidal activity by pouring 22 mL of the medium into each petriplate.
A stock solution of the pure compound was prepared at a concentration of 5 mg mL-1 in sterile dimethyl sulfoxide (DMSO). Test samples were prepared from this stock solution with dose concentrations of 100, 250, 500, and 1000 µg mL-1 by adding them to the non-solidified medium in the Petri plates. Each was marked with the respective concentration at the time when molten agar was added. Imidacloprid (Rudolf Pakistan) in powder form was used as the standard insecticide in this experiment, also at concentrations of 100, 250, 500, and 1000 µg mL-1. Ten larvae of H. armigera were placed in each Petri plate marked with the respective concentration of the fungal compound and the standard insecticide. A control treatment was maintained, consisting of only the medium and larvae. Each treatment was performed in triplicate under laboratory conditions at 30°C and 75% relative humidity.
3. RESULTS
3.1. Identification of Fungal Strains
On PDA medium, the colony color was initially white, then becoming more pinkish with the growth of the fungus over time. The colony was observed to be Fast-growing, spreading colonies that were initially circular but later became slightly irregular with age. On PDA medium, the colonies were dense but fluffy and cottony in appearance. In comparison, the same colonies on SDA and Czapec dox agar medium appeared less vibrant, more compact, and slow-growing. On the PDA medium, the colony surface was more wrinkled, specifically at the center, but on SDA and CDA medium the colony surface was smoother and even in comparison to the PDA medium (Fig. 2A). Microscopic identification was performed by observing conidia and conidiophores by staining it with lactophenol cotton blue stain and observing at 2000x using a compound microscope. The conidia were observed as small unicellular and microscopic with a dark grayish shade inside the conidiophore. The fungus under study was identified through its slenderical, erect but slightly curved shaped conidiophores which were observed both solitary and in clusters under the compound microscope (Fig. 2B).
3.2. Purification and Mass Determination of the Pure Compound
The crude extract obtained was subjected to purification via flash column chromatography, employing ethyl acetate and n-hexane as solvent systems. The elution of the pure compound was achieved utilizing a solvent system with a polarity of 85%. Subsequently, the compound underwent analysis by LCMS, employing an analytical setup comprising a Phenomenex Luna C-18 column, a PDA detector sensitive to secondary metabolites within the UV range of 200-400 nm, an autosampler, and an auto fraction collector. Following the analytical run, the pure compound exhibited a mass of 283 (Fig. 3A). For the preparative run, a total of 41.8 mg of crude extract was utilized. A concentration of 40 mg mL-1 of the crude extract in HPLC-grade methanol was prepared for the preparative run, resulting in the purification of 7.24 mg of the pure compound. The preparative process entailed a series of 37 runs each lasting for 30 minutes. The LCMS chromatogram unveiled a UV profile with a maximum wavelength (λmax) at 281 nm. The compound exhibited an electrospray (ES-) profile at 282 and an electrospray (ES+) profile at 284 [M]H+ and 306 [M]Na+, aligning with the molecular formula C14H21NO5 [calculated as C14H21NO5 + Na = 306.14] (Fig. 3B). The retention time for the compound was established at 9.6 minutes. The analysis confirmed the molecular formula of C14H21NO5 and an exact mass of 283.14 for the compound under study (Fig. 3C).
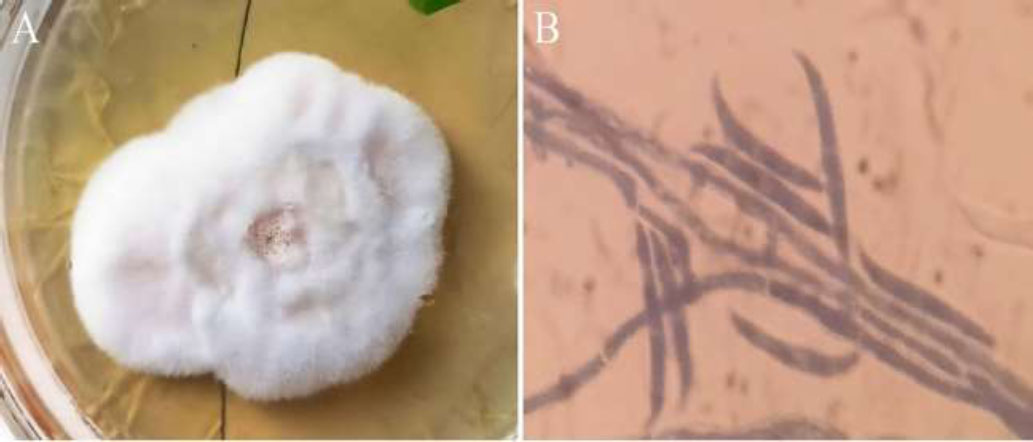
(A) Colony of F. oxysporum displaying a white-pinkish hue on Potato Dextrose Agar (PDA) medium. (B) Microscopic view showing the characteristic sickle-shaped macroconidia of F. oxysporum with cross walls.
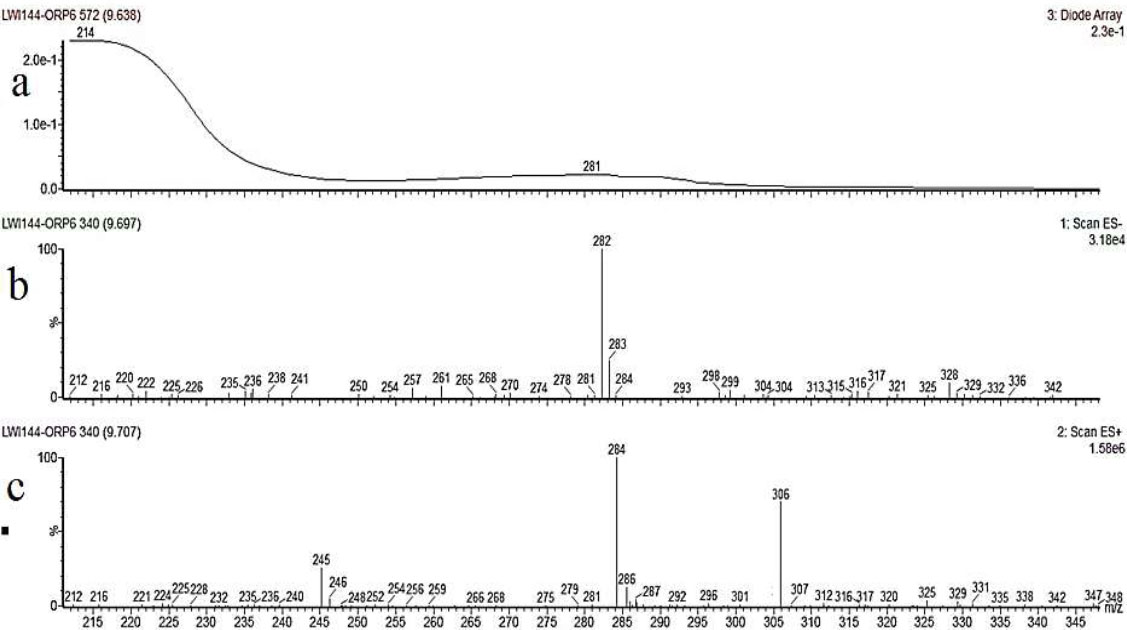
Chromatogram of the pure compound acquired from the Waters LCMS system; Line a) displays the UV absorption spectrum of the compound (281) λmax, indicating absorption at different wavelengths. Line b) represents the negative electrospray (ES-) profile of the compound, labeled as 282, while Line c) depicts the positive electrospray (ES+) profile, denoted as 284[M]H+ and 306[M]Na+.
3.3. Structure Elucidation/ Description of the Compound based on Data obtained from NMR
The structural determination of the compound was accomplished using a combination of 1H, 13C, 15N, and 17O NMR spectra, along with Heteronuclear Single Quantum Coherence (HSQC) spectroscopy. These spectra provided comprehensive insights into the different hydrogen, carbon, nitrogen, and oxygen environments within the molecule. The 1HNMR spectrum reveals multiple distinct peaks, each corresponding to different types of hydrogen environments within the molecule. The peak at δ 7.26 ppm indicates an NH proton, suggesting the presence of an amine group with strong hydrogen bonding. The peaks at δ 4.28 ppm and δ 4.11 ppm, corresponding to Atoms 2 (CH) and 10 (CH), suggest protons in relatively deshielded environments, likely adjacent to electronegative atoms or groups, such as oxygen or nitrogen. The peak at δ 6.44 ppm, corresponding to Atom 4 (CH), is indicative of a proton in an aromatic system or influenced by electron-withdrawing groups. Hydroxyl protons at δ 10.97 ppm and δ 4.19 ppm, corresponding to Atoms 13 (OH) and 15 (OH), respectively, indicate strong hydrogen bonding. The peaks at δ 2.05 ppm, δ 1.37 ppm, and δ 1.28 ppm, corresponding to Atoms 16 (CH2), 17 (CH2), and 18 (CH2), suggest these methylene groups are in an aliphatic chain. Terminal methyl groups at δ 0.97 ppm and δ 1.35 ppm, corresponding to Atoms 19 (CH3) and 20 (CH3), reflect their positions within the molecular framework and their relatively shielded environments.
The 13C NMR spectrum highlights various carbon environments within the molecule. The signal at δ 73.9 ppm for Atom 2 (CH) indicates a carbon adjacent to electronegative atoms, suggesting significant deshielding. The peaks at δ 145.1 ppm, δ 123.2 ppm, and δ 110.9 ppm, corresponding to Atoms 3 (C), 4 (CH), and 5 (C), respectively, suggest the presence of aromatic or olefinic carbons. The signals at δ 162.9 ppm and δ 159.7 ppm, corresponding to Atoms 6 (C) and 7 (C), indicate highly deshielded carbon environments, likely carbonyl groups, or similar functionalities. The peak at δ 66.1 ppm for Atom 10 (CH) suggests an aliphatic carbon adjacent to an electronegative atom. The methyl carbon signal at δ 23.7 ppm for Atom 11 (CH3) is typical for aliphatic methyl carbons. The resonance at δ 155.6 ppm for Atom 12 (C) suggests a carbonyl carbon. The signals at δ 33.8 ppm, δ 29.2 ppm, and δ 22.8 ppm for Atoms 16 (CH2), 17 (CH2), and 18 (CH2) confirm their aliphatic nature. The terminal methyl carbon signals at δ 14.0 ppm and δ 20.8 ppm for Atoms 19 (CH3) and 20 (CH3) indicate typical aliphatic environments.
The nitrogen 15N NMR spectrum provides additional insights into the oxygen environments within the compound. The peaks at δ 167.1 ppm, δ 306.0 ppm, δ 403.0 ppm, δ 248.1 ppm and δ 32.3 ppm, corresponding to Atoms 1 (O), 9 (O), 13 (OH), 14 (O), and 15 (OH), respectively, suggest the presence of various oxygen functionalities, such as hydroxyl and carbonyl groups, with the diverse chemical shifts reflecting their varied environments. The 17ONMR spectrum shows a significant peak at δ -248.4 ppm for the NH group at Atom 8, indicating a deshielded nitrogen environment, likely due to hydrogen bonding or an adjacent electronegative group. This chemical shift is consistent with nitrogen in amides or amines attached to carbonyl groups or within aromatic systems.
The HSQC spectrum provides correlations between proton and carbon atoms, confirming the assignments made from the proton and carbon NMR spectra by showing direct correlations between specific carbons and their attached hydrogens. For example, peaks in the aromatic region correlate aromatic protons with aromatic carbons, supporting the aromatic ring structure. Similarly, aliphatic protons and carbons show correlations that validate the presence of methylene and methyl groups. The spectrum highlights the detailed structure, showing which protons are bonded to which carbons, confirming the molecular framework suggested by the individual NMR spectra.
In conclusion, the integrated analysis of the 1H, 13C, 15N, and 17O NMR spectra, supported by HSQC correlations, reveals a complex organic molecule with the following features: an aromatic ring, aldehyde or conjugated systems, aliphatic chains, electronegative groups, amide or amine groups, and carbonyl and hydroxyl functionalities. This comprehensive NMR analysis reveals a molecule with aromatic characteristics, multiple functional groups, and varied carbon, nitrogen, and oxygen environments. The compound likely has a rich and complex structure, fitting the profile of many natural products with diverse functionalities. By integrating the information gathered from both spectra, the fungal compound is identified as (6-butyl-3-(1-hydroxyethyl)-2-methyl-2H-pyran-5-carbonyl) carbamic acid, revealing a complex arrangement featuring a substituted pyran ring, aliphatic chain, hydroxyl groups, and aromatic constituents (Fig. 4).
3.4. Molecular Docking Studies
The three-dimensional structure of the (6-butyl-3-(1-hydroxyethyl)-2-methyl-2H-pyran-5-carbonyl) was retrieved from Pubchem and prepared using BIOVIA Discovery Studio Visualizer. After preparation, the structure was used for docking analysis using the sigma glutathione transferase enzyme as a receptor. The docking analysis predicted -5.3 binding energy representing the selected compound has significant binding affinity for the target receptor. The docking analysis unveils that the screened compound could inhibit the target protein and block Aphis gossypii pathogenesis. Further, the docked complex confirmation analysis revealed that the selected compound has different interactions like van der Waals force, conventional hydrogen bonding, and carbon-hydrogen bonding, which are observed as different amino acids, and their interaction is presented in the following Fig. (5).
3.5. Molecular Dynamics Simulations
The static binding intermolecular conformation of the receptor and compound in docking studies was revalidated by molecular dynamics simulation assay to explore the complex dynamics on a given simulation period. The intermolecular binding mode stability of the complex was determined by two structure analyses such as root mean square deviation (RMSD) and root mean square fluctuation (RMSF), both of which were done considering carbon alpha atoms of the complex. The RMSD depicts structural changes over the simulation time and as shown in the figure, the complex revealed a very stable RMSD plot demonstrating no major structure deviations. The mean RMSD of the complex is 1.45 angstroms. Little RMSD jumps were noticed, which were observed due to flexible complex loops, and were found not to disturb the complex structure dynamics (Fig. 6). The residue level deviations were interpreted using root mean square fluctuations (RMSF) as given in Figure. The mean RMSF of the receptor in the presence of the compound is 1.58 angstroms with few sudden jumps, which corresponds to the receptor loops due to small compound pocket movements (Fig. 7).
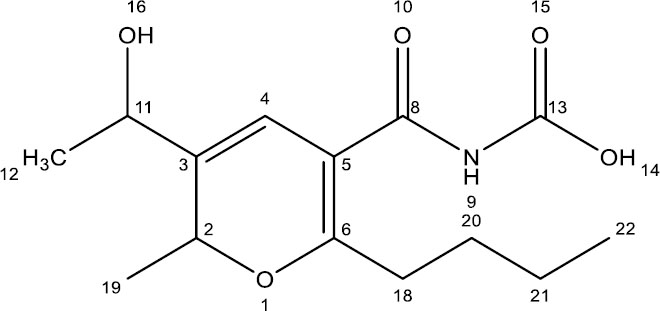
Structure of the pure compound.
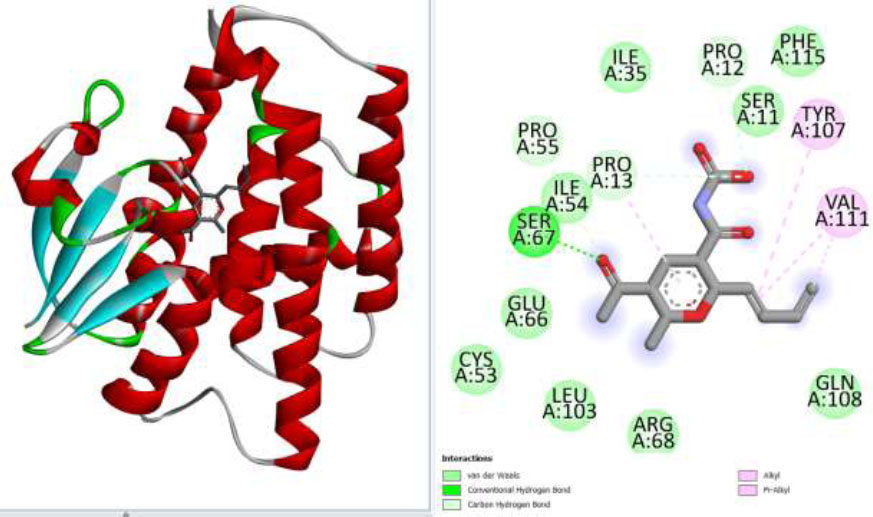
(A) Represent the selected compound docked with targeted protein while (B) represents different binding interactions of compound (4-chloro-1-(methyl)- λ4-sulfaneyl)-1 λ5-phosphinine-3-carbonyl) carbamic acid) and selected proteins (sigma glutathione transferase).
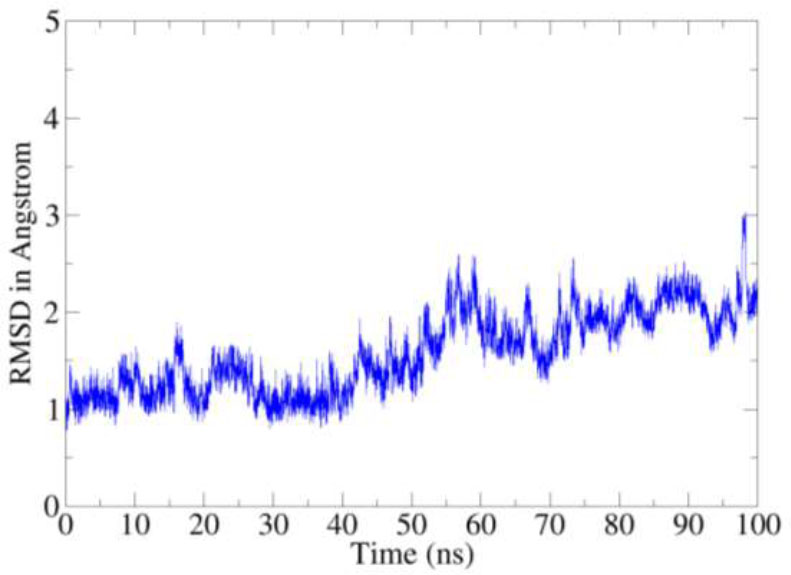
RMSD based on carbon alpha atoms.
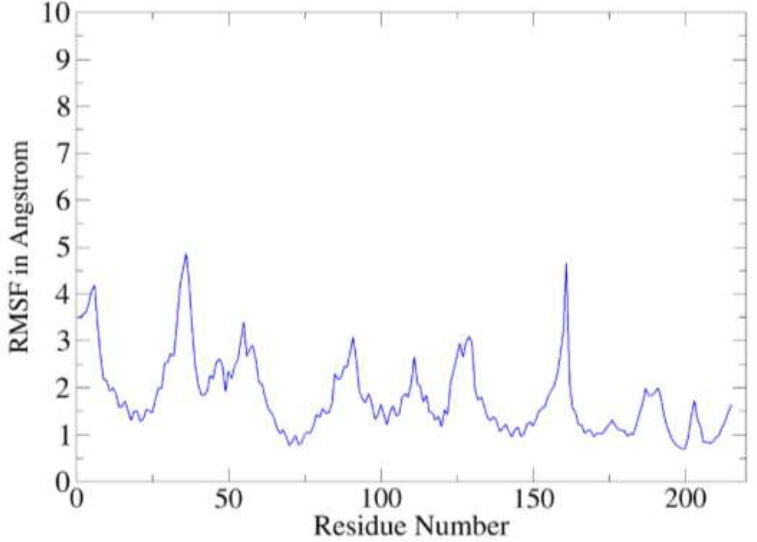
RMSF analysis of the complex.
3.6. Binding Free Energies
The binding free energies for the compound in complex with the receptor enzyme were determined along the simulation trajectories. It was found that the intermolecular binding affinity was dominated by van der Waals forces, followed by electrostatic forces, while a negative contribution was seen from the solvation energy. The van der Waals, electrostatic, and solvation energy of the complex was -68.64 kcal/mol, -25.75 kcal/mol, and 10.58 kcal/mol, respectively. The total energy contribution of the complex was -83.81 kcal/mol, illustrating the formation of a highly stable complex. The Table shows details of the different energy contributions to the complex formation (Table 1).
| Parameter | Compound 283 |
|---|---|
| Energy van der Waals | -68.64 |
| Energy Electrostatic | -25.75 |
| Total Gas Phase Energy | - 94.39 |
| Total Solvation Energy | 10.58 |
| Net Energy | -83.81 |
3.7. In vitro Insecticidal Activity of the Compound
This experiment was conducted in triplicate, and the mean of the three trials was considered in the compilation
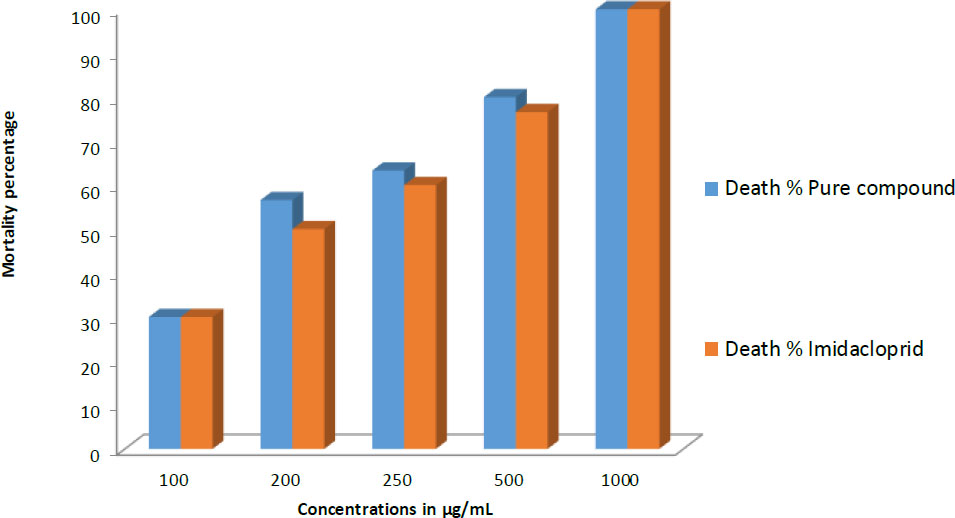
Comparison of insecticidal activity of the purified fungal compound and the standard insecticide (Imidacloprid) against Helicoverpa armigera larvae at different concentrations. The Y-axis represents the mortality percentage of larvae, while the X-axis indicates the tested concentrations. The blue bars correspond to the mortality percentage induced by the pure compound, while the red bars represent the mortality percentage caused by Imidacloprid.
of results. The fungal compound, administered at concentrations of 100, 200, and 250 µg mL-1, exhibited mortality rates of 30%, 56.6%, and 63.3%, respectively. Specifically, at the 100 µg mL-1 concentration, it resulted in the death of 3 larvae in each Petri plate. At the 200 µg mL-1 concentration, 6, 6, and 5 larvae succumbed in the corresponding Petri plates, while at the 250 µg mL-1 concentration, 6, 6, and 7 larvae were killed. In comparison, the standard drug, imidacloprid, demonstrated mortality rates of 30%, 50%, and 60% at the respective concentrations. At a concentration of 500 µg mL-1, the fungal compound inhibited the growth of 8 larvae in each test plate, resulting in an 80% mortality rate. Meanwhile, the standard at this concentration caused the death of 8 larvae in two Petri plates and 7 larvae in the third, resulting in a mortality rate of 76.6%. At the highest applied concentration (1000 µg mL-1), both the fungal compound and the standard exhibited 100% mortality by inhibiting the growth of every larva in all Petri plates. The results of in vitro insecticidal activity are summarized in Fig. (8).
4. DISCUSSION
Endophytic fungi are a prolific source of secondary metabolites with diverse applications, including antimicrobial, anticancer, and insecticidal properties. Commercial insecticides derived from fungal sources, such as Mcotrol [42], PFR-97 [43], and Met52 [44], highlight the significant potential of these natural products for effective pest management in agriculture. While the genera Beauveria, Metarhizium, and Isaria have been extensively studied for their insecticidal metabolites [45], Fusarium species, such as Fusarium oxysporum and Fusarium solani have also been recognized for producing notable insecticidal compounds [46]. In the present study, we isolated F. oxysporum from the rhizosphere of ginger plants and purified four secondary metabolites. This study specifically concentrates on one of these purified compounds. The compound's structure was elucidated, and its potential as an insecticide was explored through comparative structural analysis, in silico, and in vitro studies. Our findings suggest that this compound could serve as an effective insecticide in agricultural settings, aligning with previous reports of Fusarium species producing insecticidal metabolites.
The structural features of the pure compound, 6-butyl-3-(1-hydroxyethyl)-2-methyl-2H-pyran-5-carbonyl carbamic acid, were compared to established insecticides, revealing several intriguing parallels. Insecticides from various chemical classes, including organophosphates, pyrethroids, neonicotinoids, and carbamates, function through diverse mechanisms targeting critical physiological processes in insects. The presence of a carbamate moiety in our compound suggests a mode of action similar to that of carbamate insecticides, which inhibit acetylcholinesterase, disrupting neural signaling in insects [47]. This structural resemblance implies potential neurotoxic effects akin to traditional carbamate insecticides. Furthermore, the substituted pyran ring in our compound evokes structural similarities with pyrethroid insecticides, known for their heterocyclic structures. Pyrethroids act by prolonging the opening of voltage-gated sodium channels in insect neurons, leading to hyperexcitation and paralysis [48]. The analogous structure suggests a comparable mechanism of action for our compound. Additionally, the aliphatic chain in the compound is reminiscent of the hydrophobic alkyl chains in synthetic pyrethroids, which enhance cuticular penetration and binding affinity to insect nerve membranes, increasing insecticidal potency [48].
The hydroxyl group within our compound bears a resemblance to functional groups found in neonicotinoid insecticides. Neonicotinoids act as agonists at nicotinic acetylcholine receptors in insects, disrupting neurotransmission and ultimately leading to paralysis and death [49]. This structural feature suggests potential interactions with similar target sites, implying a comparable mode of action. Based on the structural features of the purified compound, we hypothesized that the compound under study possesses inherent insecticidal properties. This hypothesis was experimentally validated through in silico and in vitro analyses, comparing the compound's efficacy with a standard insecticide, Imidacloprid.
The compound's in vitro insecticidal potential was evaluated against H. armigera. At the highest applied concentration (1000 µg mL−1), both the fungal compound and Imidacloprid exhibited 100% mortality. However, at lower concentrations, the fungal compound demonstrated superior efficacy. Specifically, at concentrations of 100, 200, and 250 µg mL−1, the fungal compound achieved mortality rates of 30%, 56.6%, and 63.3%, respectively, compared to Imidacloprid's 30%, 50%, and 60%. At a concentration of 500 µg mL−1, the fungal compound resulted in an 80% mortality rate, compared to 76.6% for Imidacloprid. These results indicate that the fungal compound possesses significant insecticidal activity, even at lower concentrations. This feature is ecologically significant, as it allows for the use of lower concentrations of the insecticide to effectively control insect populations in agricultural settings. This approach potentially reduces environmental impact and the risk of developing resistance.
Our findings highlight the potential of the compound under study isolated from F. oxysporum as an effective insecticide. The superior efficacy at lower concentrations suggests it could be a valuable addition to integrated pest management strategies. However, further investigations into its precise mode of action, environmental impact, and field efficacy are necessary to fully understand its potential for sustainable agricultural pest management. Future studies should also explore the synthesis of analogs to optimize their insecticidal properties and assess their compatibility with other pest control methods. This preliminary investigation provides evidence for the insecticidal potential of the purified compound from F. oxysporum. The structural and experimental analyses offer insights into its mechanisms of action, supporting its candidacy as a potent insecticide. This work paves the way for developing new, ecologically friendly pest control solutions derived from fungal secondary metabolites.
CONCLUSION
Based on the results obtained from the in silico and in vitro experiments, it is evident that the pure compound under study exhibits potent insecticidal potential. Molecular docking studies demonstrated a significant binding affinity for sigma glutathione transferase, with a binding energy of -5.3 kcal mol-1, indicating strong inhibitory potential. Molecular dynamics simulations confirmed the stability of the compound-receptor complex, supported by consistent RMSD and RMSF values. The binding free energy analysis revealed that van der Waals forces predominantly drive the interaction, resulting in a net binding energy of -83.81 kcal mol-1, suggesting a highly stable complex. Furthermore, the in vitro assay showed substantial larvicidal activity, with mortality rates reaching 100% at the highest concentration. From these collective findings, we conclude that this F. oxysporum-derived compound is a promising candidate for insecticide development and warrants further investigation for practical pest control applications.
STUDY LIMITATION
This study serves as a preliminary investigation into the purification, characterization, structural elucidation, and potential applications of a bioactive compound derived from Fusarium species. Our primary objective was to explore its potential using in silico and in vitro analyses. However, certain limitations should be acknowledged.
The compound was isolated in a limited quantity, restricting us to conducting only a single in vitro bioassay. Due to this constraint, further biological evaluations, including comprehensive toxicity profiling and field trials, could not be performed at this stage. The small yield also limited detailed investigations into the compound’s mechanism of action, metabolic stability, and selectivity, particularly regarding its potential effects on non-target organisms, including humans. Additionally, field trials, which are essential for assessing the compound’s real-world insecticidal efficacy, could not be conducted. Despite these limitations, this study establishes a foundation for future research, emphasizing the need for chemical synthesis of the compound in larger quantities to enable further toxicity assessments, field applications, and environmental impact studies.
PROSPECTS
This study lays the groundwork for further research into the bioactive potential of the purified Fusarium-derived compound, particularly as a promising eco-friendly insecticide. Future work will focus on chemical synthesis to produce the compound in sufficient quantities for comprehensive biological evaluations, including detailed toxicity profiling, selectivity assessments, and environmental safety studies. Additionally, field trials will be conducted to evaluate its real-world efficacy in pest management. Further investigations will explore its broader bioactive properties, such as antimicrobial, antifungal, or plant-growth-promoting activities, which could expand its potential applications in agriculture and biotechnology.
AUTHORS’ CONTRIBUTIONS
The authors confirm their contribution to the paper as follows: S.K.: Study conception and design; M.S.: Data collection; A.M.A.: Validation; S.A., D.Q.W., N.A.A.: Draft manuscript. All authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| PDA | = Potato Dextrose Agar |
| CDA | = Czapec Dox Agar |
| SDA | = Saboraud Dextrose Agar |
| HCl | = Hydrochloric acid |
| LCMS | = Liquid Chromatography-mass Spectrometry |
| RMSD | = Root Mean Square Deviation |
AVAILABILITY OF DATA AND MATERIALS
All data generated or analyzed during this study are included in this published article.
FUNDING
King Saud University, Riyadh, Saudi Arabia, has funded the study under the project number (RSPD2025R1035).
ACKNOWLEDGEMENTS
The authors are thankful to the Researchers Supporting Project number (RSPD2025R1035), King Saud University, Riyadh, Saudi Arabia.


