All published articles of this journal are available on ScienceDirect.
Next-generation Sequencing in Newborn Screening: A Review on Clinical and Economic Prospects
Abstract
Next-generation sequencing (NGS) technology has revamped the area of genetics with improve the sequence of a substantial number of genes with high accuracy and short turn-around time. It may allow being used of NGS as a first-tier diagnostic test for inborn errors of metabolic or other genetic disorders. Early diagnosis of genetic disorders may help to improve the clinical condition of the child. The advantages of NGS included panel-specific gene sequencing, which targets disease-specific genes to confirm the genetic conditions. This review discussed the advantage and potential challenges of the NGS in newborn screening, other methodologies for newborn screening v/s NGS. The application of NGS in various disorders, in comparison to the clinical importance, and economic aspects of other existing technologies are also discussed. Gene-specific panels and whole exome sequencing have shortened the clinical diagnosis of complex medical conditions at an early age. Furthermore, gene sequencing facilitates to recognize the novel mutations. There are innumerable gaps in between knowledge, as well as the views of varied populations, abilities of public health, and health economics. DNA sequencing through NGS is nowadays frequently used in some clinical diagnoses, and its execution in newborn screening can provide us with better outcomes. Although inferences across the countries additional rigorous cost-effectiveness studies towards NGS have to be piloted and it is a favour to use NGS for newborn screening. In conclusion, NGS is a rapid, robust, and accurate diagnostic tool that can be used for newborn screening which helps the clinician to make a correct diagnosis and help in prior prevention and surveillance of disorder conditions.
1. INTRODUCTION
Next-generation sequencing (NGS) has the potential to improve the diagnostic and prognostic success of newborn screening (NBS) programs [1]. It is an effective tool for identifying various genetic alterations in newborns. NGS has transformed the field of genetics, providing the ability to sequence a substantial number of genes within a short turnaround time in a multiplexing manner [2]. The utility of NGS has dramatically increased, which may be suggestive of the particular disorder with noteworthy cost decreases and wider community acceptance [3]. As a direct result, the ability to make inherited disorders diagnoses has increased via NGS. It can be performed using dried blood spot (DBS) of neonates through isolation of DNA and has the prospective to improve the investigative and prognostic efficiency of NBS programs [4]. However, a few more demanding clinical-effectiveness studies related to NGS have to be conducted on NBS, which are required to define whether NGS can increase screening results in terms of sensitivity and specificity [5]. Several government health organizations were recognized to confirm that NBS should be standardized and uniform [6]. Since then, NBS has continued to explore the best framework moving forward as technology diagnosing children with genetic conditions continues to emerge [7]. Furthermore, there are various gaps in between knowledge, as well as the views of varied populations, abilities of public health, and clinical effectiveness inferences [8]. Keeping the following view, this article with highlights the current advancements in the usage of NGS in NBS, the obstructions in its application in NBS, and the extensive impacts of its use in the NBS program. The main objective of NGS in NBS is to be the targeted investigation and distinction of gene variants considering a suspect of preventable or curable conditions [9]. Fig. (1) summarizes the brief overview of NBS through NGS, NBS challenges, and opportunities for population health. In addition, this article provide a brief history and outline of the current state of NGS regarding NBS including recent recommendations and prospective tasks for the usage of such tools in NBS.
2. NEWBORN SCREENING FOR VARIOUS GENETIC DISORDERS
NBS program was introduced by the World Health Organization (WHO) as per Wilson and Jungner guidelines for the development and implementation of a screening test [10]. Although the tests supplementary provided the outline for the additional development and amendment of NBS [11]. It is a public health program, which aims to identify pediatric populations with medically unfit conditions earlier than becoming symptomatic [12]. Over the past few decades, the number of medical conditions in children has gradually increased. These medical conditions are auxiliary to the screening program in different areas across the world [13]. The NBS program interventions can provide long-term results in terms of improvement and carrier identification [14]. The program was underway with identifying phenylketonuria (PKU), hemoglobinopathies to a diagnosis of sickle cell disease, glucose-6-phosphate dehydrogenase (G6PD) deficiency, congenital hypothyroidism (CH), congenital adrenalhyperplasia (CAH), tyrosinemia type I (HT1), cystic fibrosis (CF), etc [15]. The diagnosis and exploration of these disorders are challenging and time-consuming due to the rarity of the individual disorder, the genetic heterogeneity, and variability of the clinical manifestations, and the lack of laboratory testing [16]. Various NBS programs have been experienced in frequent countries since the start of NBS [17]. NBS was started in the 1960s and phenylketonuria screening was successfully implemented by different nations across the world first [12].
3. NEWER TECHNIQUES TO EXPAND NEWBORN SCREENING PROGRAM
Several health organizations were recognized to confirm that NBS should be standardized and uniform. Since, the field of NBS has continued to explore the best framework moving forward as technology for diagnosing pediatric populations with genetic conditions continues to emerge [8, 18, 19]. With the progress and the availability of electrospray ionization (ESI) tandem mass spectrometry (MS/MS), the instrument can measure various metabolites related to inborn errors of metabolism (IEM) disorders [20]. It was the first great extension of the NBS programs across the world. Furthermore, future development for the screening of rare disorders also needs to be required, which needs additional diagnostic tests [21]. To facilitate more accuracy, a combined screening test can be used MS/MS and targeted-NGS in a parallel way, which may be more useful to offer the potential to enhance performance [22]. It can increase the specificity results by identifying new disease-associated variants along with specifying genotypes to identify the carriers [23]. Fig. (2) summarizes the intermittent techniques used individually or combinedly for the NBS.
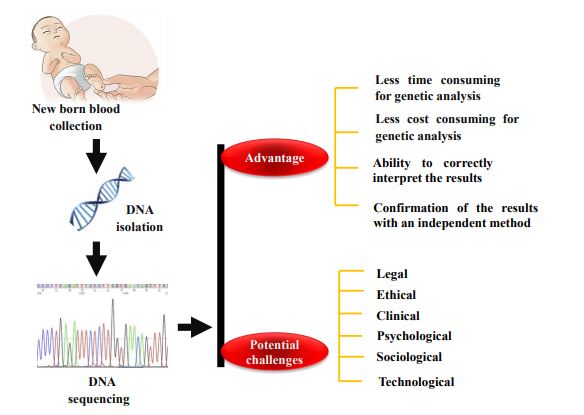
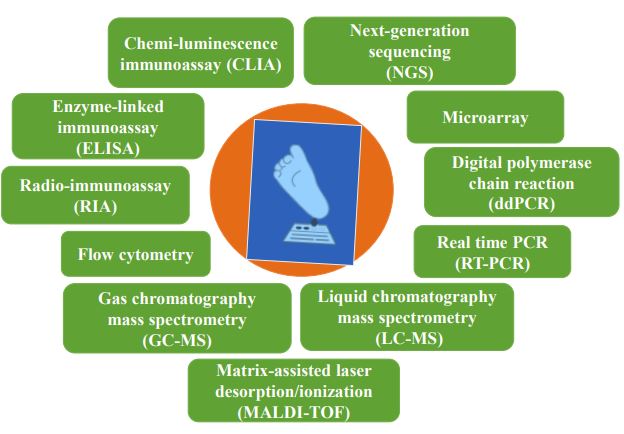
4. NEXT-GENERATION SEQUENCING IN NEWBORN SCREENING PROGRAM
NGS has cutting-edge technology and transformed the field of genetics using molecular biology, providing the ability to sequence a large number of important genes within a short turnaround time [24]. As a direct result, the ability to make genetic diagnoses has increased powerfully [25]. Genome sequencing is now often used in various medical diagnoses, and its implementation in NBS has also been extensively encouraged [26]. In the last decade, there is a significant advancement toward high-throughput DNA sequencing, which resulted in a reduction in both the cost and time required for genetic analysis [1, 9]. These methods are widely used in massive multi-parallel sequencing. The NGS can investigate all protein-coding genes or even entire genomes in one single time via whole-exome sequencing (WES) or whole-genome sequencing (WGS) [27]. The ease of accessibility and a considerable decrease in costs of DNA sequencing has raised the ultimatum of whether metabolic NBS should be replaced by WES/WGS [8]. While NGS can potentially identify the number of disorders and the generalization of its practice raises many important ethical issues [28]. These developments can result in an escalation of genetic diagnoses in a shorter time [29]. Fig. (3) summarizes the common recommended diseases for NBS and their targeted genes.
Despite the health benefit of genetic testing in the clinical setting, the testing in asymptomatic children has been widely questioned [30]. Given the limited knowledge of rare genetic conditions, large prospective studies of healthy individuals should be incorporated in the decision to include conditions on an NGS-NBS panel, especially in circumstances with no current actionable treatment [26]. Genomic information can help manage health throughout life and can be applied to thoughtful children [23, 31]. Nevertheless, the current research focusing on long-term outcomes and psychological impact is needed before applying the technology to population screening [32]. Milko et al. piloted the NBS through NGS, and the Newborn Exome Sequencing for Universal Screening (NC NEXUS) was conducted to assess the technical feasibility and limitations of NGS-NBS. Furthermore, it was planned and evaluated framework to convey various types of genetic information [33]. Recently, NeoSeq, an economic genomic screening test for NBS utilized for the most inborn errors of metabolism, which can reduce the rate of false positive results, shorten the porting cycles, and reduce the screening cost also [22, 34]. Genetic disorders are the foremost reason for mortality and morbidity in newborns in neonatal and pediatric intensive care units (NICU/PICU) [35]. One study reported that rapid whole-genome sequencing (rWGS) increased the proportion of NICU/PICU infants receiving a genetic diagnosis within 28 days [36]. In China, Children’s hospitals developed a panel of 465 causative genes for 596 early-onset, relatively high incidence, and potentially actionable severe inherited diseases in NBS with the Targeted Sequencing (NESTS) program and suggest that NESTS is feasible and cost-effective as a first-tier NBS program, which can change the status of current clinical practice of NBS in China [37]. However, it is still necessary to further optimize the panel design and add more clinically relevant genomic variants to increase its sensiti-
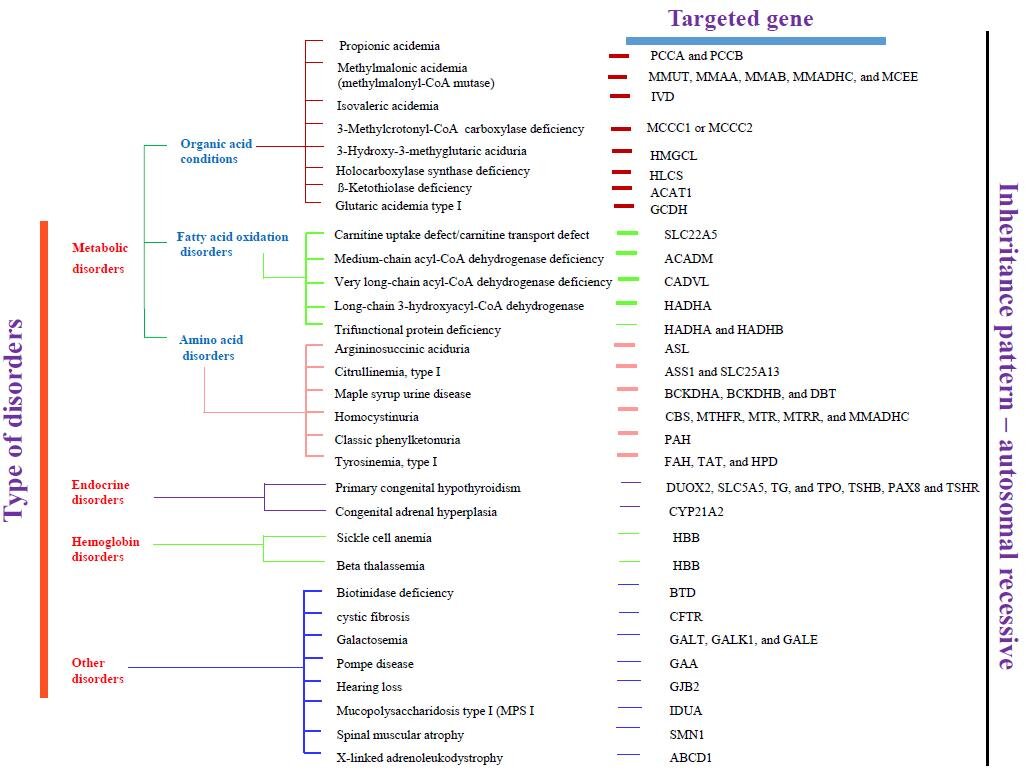
vity [22]. Boemer et al. 2017 reported that NGS using dried blood spots can detect causal mutations in patients with inherited metabolic disorders [38]. The NGS-based NBS approach is idyllic for the identification of newborns with monogenic forms of IEM, which are heterogeneous but can be easily distinguished [39]. The NGS with the amplicon-based panel has also been validated for diagnostic purposes and can be used for second-tier NBS IEM tests [40].
5. NEXT-GENERATION SEQUENCING ON THE WAY TO CLINICAL AND ECONOMIC VIEWPOINTS IN THE NEWBORN SCREENING PROGRAM
The extension of NBS has raised many clinical, ethical, lawful, psychological, sociological, and scientific distresses over time [1]. As the cost of NGS continues to decrease and our understanding of genomic databases continues to improve, demands have been elevated to apply NGS on a larger scale with improved outcomes for potential health concerns [41]. There are various gaps in between facts, as well as the views of diverse populations, capabilities of health care systems, and health economic implications. It will be essential to scrupulously evaluate the outcomes and ensure the plans can evolve to maximize benefit. The ease of accessibility and substantial decrease in costs of DNA sequencing has raised the question of whether metabolic NBS should be replaced by WGS/WES [42]. While in newborn genome sequencing can potentially increase the number of disorders identified and the simplification of its practice raises some important ethical issues [28]. The development of the map of the human genome assured a new era in medicine [43]. In the last few decades, the usefulness of DNA sequencing for health benefits has been demonstrated during manifold periods and is now a routine in some areas of medicine [43]. Advanced DNA sequencing techniques and truncated cost offered the promise of a personalized genome to provide a personal health benefit [44]. The first usage of genome sequencing for disease diagnosis befell in 2009 at the University of Wisconsin for a child with severe inflammatory bowel disease. The cause of the disease is the occurrence of the pathogenic variant of the XIAP gene [45]. The newborn screening cost via the traditional method in Europe is 1 to 44 Euro (€1: screening for 2 conditions; €43.24: screening for 17 conditions) and in the United States is around 100 USD ($100: screening for 30 conditions) per test [1]. Few studies should be commenced to obtain accurate data about all costs involved as part of NBS including subsequent diagnostic costs as well as the costs of not implementing NGS. The price of NGS for million bases (i.e. 1 Mb) has considerably fallen and may continue to fall even extra. However, the cost of NGS is still high other than the existing NBS programs. The cost of NGS in NBS, pre-and post-testing, is also a key issue in the decision to implement NGS in NBS. Furthermore, the setup and human resource costs to ensure monitoring, appropriate education, counselling, interventions, and storage needs to be assessed [9, 30].
CONCLUSION AND FUTURE PROSPECTS
NGS is a rapid, robust, and accurate diagnostic tool that can be used for newborn screening, which helps the clinician to make a correct diagnosis and help in prior prevention and surveillance of disorder conditions. It is of substantial interest whether NGS is applicable for NBS and has great potential to improve NBS through the understanding of genetics, which can provide flawless diagnosis. The expansion of the NBS program for various disorders needs to be required, which calls for additional tests. Necessary characteristics of a screening test include the availability of an economical method of identification of important health problems for which treatment is available, as well as the infrastructure to provide such treatment. It can sequence all protein-coding regions or even entire genomes at one single time, resulting in an increase in genetic diagnoses and a shortened time to diagnosis for patients with expected genetic disorders. A broader understanding of several aspects is needed to facilitate the incorporation of NGS into newborn public health screening programs and inform the development of important future NBS policy guidelines.
LIST OF ABBREVIATIONS
| DBS | = Dried blood spot |
| IEM | = Inborn errors of metabolism |
| MS/MS | = Tandem mass spectrometry |
| NBS | = Newborn screening |
| NGS | = Next-generation sequencing |
| WES | = Whole exome sequencing |
| WGS | = Whole genome sequencing |
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
Dr. Amit Kumar Mittal is an Editorial Advisory Board of The Open Biotechnology Journal.
ACKNOWLEDGEMENTS
The authors AKM and VJ would like to thank the Department of Health Research (DHR) and author DSS would like to thank the UMMID initiative, the Department of Biotechnology (DBT), Govt. of India for the financial support.


