All published articles of this journal are available on ScienceDirect.
Expression and Characterization of Rice-produced Recombinant Porcine Lactoferrin and its Antioxidant Activities
Abstract
Background:
Lactoferrin (LF) exhibits multiple beneficial biological activities and thus has been used as a health food and additive. To broaden its application in the food industry, the porcine LF (pLF) gene has been engineered into rice to produce recombinant LF (rpLF) for use as a food additive. The iron-binding and antimicrobial activities of rpLF and its positive effects on early weaned piglets have been previously evaluated, yet several features, such as the signal peptide removal, glycosylation sites and antioxidant activity of rpLF, have not been fully characterized.
Objective:
In this work, the rice-produced rpLF was purified and its biochemical structure and antioxidant activities characterized.
Methods:
HPLC, Western blot, PAS/VVL/PNA staining, Edman degradation assay, MALDI-TOF, LC-MS/MS and antioxidant activity assays were performed.
Results:
The results showed that this purified rpLF is a mature form of LF; its signal peptide was correctly removed, and two N-glycosylation sites located at N365 and N472 were identified. The molecular mass heterogeneity of rpLF could be eliminated by treatment with PNGase glycosidase, suggesting that different degrees of N-glycosylation occur in rpLF. A series of assays including the iron chelating activity, reducing power assay, lipid peroxidase activity and radical-scavenging activity showed that the antioxidant activity of rice-produced rpLF was equivalent to that of bovine LF.
Conclusion:
Rice-produced rpLF was correctly processed post-translationally and displayed antioxidant activity equivalent to that of bovine LF; thus, rice-produced rpLF can be recognized as a plant-based antioxidant to be used as a functional additive in animal feed and for the food industry.
1. INTRODUCTION
Lactoferrin (LF) is an iron-binding glycoprotein of the transferrin family and was first discovered in bovine milk; LF proteins isolated as well as characterized from the milk of various mammals exhibit high amino acid sequence homology [1]. LF is produced by epithelial cells and can be found in high concentrations in milk and exocrine secretions such as tears, saliva, semen, cervical mucus and sweat [2, 3]. LF has multiple functions, including modulation of both the inflammation and immune systems, as well as activity against bacteria, fungi, RNA and DNA viruses, and some parasites with iron-binding and structural properties [4-9]. The primary structure of LF contains various Cys residues that allow the formation of intramolecular disulfide bridges and Asn residues in the N- and C-terminal lobes that provide potential N-glycosylation sites to which complex and high-mannose-type glycans are linked [10, 11]. These glycans are structurally heterogeneous, resulting in various molecular mass isoforms of LFs, and this heterogeneity may be responsible for various biological functions [10-12].
Glycosylation is a type of post-translational modification in eukaryotic and prokaryotic cells that leads to the formation of various functional glycoprotein complexes [13, 14]. N-linked glycosylation involves the addition of glycans to asparagine (Asn) residues of a protein at the highly conserved Asn-X-Ser/Thr motif, and as a glycoprotein, LF is N-glycosylated at the conserved Asn-X-Ser/Thr motif, but various glycans are added at different glycosylation sites [10-12, 15]. For example, bovine LF (bLF) has five conserved N-glycosylation motifs, Asn233, Asn281, Asn368, Asn476, and Asn545, but only four sites (Asn233, Asn368, Asn476, and Asn545) are commonly glycosylated [16]. Human LF (hLF) showed three conserved N-glycosylation sites, Asn138, Asn479, and Asn624, and only two sites (Asn138 and Asn479) are commonly glycosylated [17]. Recombinant hLF (rhLF) expression in transgenic cattle appears to be glycosylated at the same two sites [15], as well as rice-produced rhLF [18], although various N-linked glycan structures have been observed. Therefore, the selected N-glycosylation sites of LF are likely conserved between its animal sources and its transgenic recombinants. In porcine LF (pLF), two potential N-glycosylation sites located at N365 and N472 (Fig. S1-A) have been proposed [19]; however, experimental demonstration has not been reported. In the present study, an MS/MS assay was applied to determine the N-glycosylation sites of rice-produced rpLF.
Given that only a limited number of animal colostrum contain high concentrations of LF, numerous attempts to obtain large amounts of recombinant LF using yeast [20, 21], bovine [22, 23] and plants [24] have been implemented. Among these various protein production systems, plant-based recombinant LF has the advantages of potential production scalability and cost reduction and thus has become an attractive alternative option [24]. The expression of recombinant LF in plants, such as tobacco [25], potato [26], sweet potato [27], wheat [28], barley [29], alfalfa [30] and rice [31-33], has been established. To ensure high expression of transgenes in plants, factors that affect transcription, translation, and post-translational modifications at various levels have been evaluated and reviewed [34]. The strength and expression from the promoter, a key regulatory element, play important roles in achieving high levels of transcription. The promoters of the cauliflower mosaic virus 35S and rice actin genes have been characterized, and studies showed that these promoters are efficient in regulating the constitutive expression of a foreign gene in transgenic rice (reviewed by Ignacimuthu et al., 2000) [35]. In addition, matrix attachment region (MAR) sequences, which could ensure promoter activity and enhance gene expression in the genome, have been incorporated into vector constructs and flanked with target genes to stabilize and increase transgene expression in transgenic plants [36-40]. In the present study, vectors containing a 35S promoter, rice actin promoter, or a rice actin promoter with an MAR sequence were created, and their ability to drive the expression of the porcine lactoferrin gene in transgenic rice was determined.
In addition to the advantages of recombinant protein production scalability and cost reduction, plant-produced recombinant LFs retain the same biochemical and physiological properties as their native counterparts and can be used as pharmaceuticals or food additives [18, 24, 31-33, 41]. For example, a rice-derived recombinant porcine LF (rpLF) improved the growth and immune characteristics of early weaned piglets when used as a food supplement [31]. Although several biological activities of these plant-produced recombinant LFs, including antimicrobial activity, have been evaluated [31-33, 41, 42], studies on their antioxidant activity have not yet been thoroughly investigated.
Oxidative damage is considered a metabolic disturbance that affects physiological processes, and its presence affects the health of animals [43]. Antioxidants that can reduce oxidative damage can be of great benefit in improving the quality of life by preventing or postponing the onset of degenerative diseases. Therefore, the search for antioxidants has always been an attractive field of research. Bovine LF, with iron-binding ability, was demonstrated to possess antioxidant activity [44] and has been considered an antioxidant [45-47], and LFs from other animal sources, such as humans [48] and camels [49], exhibited antioxidant activity as well. However, the activity of native lactoferrin cannot be automatically assumed to be identical to its recombinant product from transgenic plants. Whether recombinant LF produced from transgenic plants retain their antioxidant activity has not yet been fully evaluated; thus, demonstration of the antioxidant activity of plant-based recombinant LFs would be beneficial.
Antioxidants are classified into four types, including metal chelators, chain breakers, peroxide decomposers and reactive oxygen scavengers, which have different mechanisms of action. Among various metals, ferrous ions have been considered the oxidation catalysts that most frequently cause the acceleration of lipid oxidation. Therefore, the ferrous ion-chelating ability is usually used as an antioxidant assessment index [50]. Furthermore, DPPH (1,1-diphenyl-2-picrylhydrazyl) is a relatively stable artificial free radical; it is often used to evaluate the free radical scavenging ability of an antioxidant by measuring the absorbance value to determine the amount reduced by the antioxidant. Various assays with different methods are available for investigating the antioxidant properties of samples of interest [51]. Four methods, including 1) metal chelating capacity assay, 2) lipid peroxidation inhibition assay, 3) reducing power assay and 4) free radical scavenging capacity assay, were chosen and performed to determine the antioxidant activity of rpLF.
2. MATERIALS AND METHODS
2.1. Materials
The materials ascorbic acid (Vit C), butylated hydroxytoluene (BHT), bovine lactoferrin (bLF) and EDTA used for antioxidant activity assays were purchased from Sigma (MO, USA).
2.2. Plasmid Vector Construction
A full-length cDNA fragment (2061 bp) of porcine lactoferrin (accession number NM 214362) driven by a rice actin promoter (1255 bp) [52] and Nicotiana tabacum RB7 MAR DNA fragment (accession number U67919) were first constructed in pUC18 and then inserted into the HindIII site of the vector pCAMBIA1300 (accession number AF234296) to form pCAM-Act-MLF (Fig. 1). This plasmid vector was then transferred to Agrobacterium tumefaciens strain EHA105 for rice transformation. The hygromycin phosphotransferase (hpt) gene driven by the 35S promoter was used as a selection marker.
2.3. Preparation of Porcine rLF from Rice Grains
One hundred grams of mature rice grains from transgenic rice were ground in liquid nitrogen in extraction buffer (20 mM Tris/HCl, 1 M NaCl, and 0.1% (v/v) Triton X-100, pH 7.4). The extracted proteins were partially purified by 4-55% ammonium sulfate precipitation; the precipitated proteins were collected by centrifugation and then resuspended and dialyzed against the initial buffer to remove excess salts. These proteins were further purified through a heparin-Sepharose column with an AKTA purifier 10 system (Amersham Pharmacia Biotech, Inc., UK). The amounts of purified protein were detected by the Bradford method using a protein assay kit (Bio-Rad, USA).
2.4. Western Blot Assay
The antibody to be used against rpLF was raised in a rabbit, and an anti-LF antiserum was prepared and purified as described by Wu et al. [53], where its specificity against both the E.coli-expressed and rice-produced rpLFs had been evaluated (Fig. S2). Proteins from whole grains of TNG67 and rice bran as well as the whole grains and leaves of transgenic lines were extracted as described by Lee et al. [31]. The protein samples were then fractionated on a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane with a BioRad Trans-Blot system according to the manufacturer’s instructions. The membrane was first probed with primary anti-LF antibody and then treated with secondary anti-rabbit antibody conjugated with alkaline phosphatase (Sigma, MO, USA). After antibody recognition, the membrane was incubated with NBT (nitro blue tetrazolium chloride, 50 mg/mL) and BCIP (5-bromo-4-chloro-3-indolyphosphate, 50 mg/mL) in 10 mL of alkaline phosphatase buffer (100 mM NaCl, 5 mM MgCl2, and 100 mM Tris-HCl, pH 9.5) for color development [54].
2.5. Enzyme-linked Immunosorbent Assay
The microtiter plates were coated with 50 μl of protein overnight at 4°C. After blocking with 2% nonfat milk in PBS for 2 hours and rinsing three times with PBS, the rpLF antibody (50 μl) was added to each well and incubated for 2 hours. Then, 50 μl of alkaline phosphatase conjugated anti-rabbit IgG (Sigma-Aldrich, USA), the secondary antibody, was added. The reactions were then incubated with p-nitrophenyl phosphate substrate (Sigma, MO, USA) in diethanolamine buffer (pH 9.8) for 1 hour in the dark at room temperature. The absorbance at 405 nm was read in a Bio-Kinetic Reader EL 312e (Bio-Tek Instrument Inc., VT, USA).
2.6. Determination of Molecular Weight and N-terminal Sequence
A MALDI-TOF-MS instrument mode voyager ED PRO (Applied Biosystems, USA) equipped with a nitrogen laser radiating at 337 nm was used to obtain protein mass spectra. The MALDI matrix was prepared as a saturated solution of SA (sinapinic acid) in 50% acetonitrile containing 0.1% TFA. One microliter of the mixture was spotted on the sample plate and allowed to dry. The spectra were acquired in linear mode at an acceleration voltage of 25 kV, 90% grid voltage, 0.15% guide wire voltage, and 750 nsec delay with external calibration. The spectrum was obtained by accumulating an average of 200 shots in the positive ion mode. The N-terminal sequence of desalted protein samples was analyzed by SDS-PAGE following a reported protocol and sequenced using a Procise protein/peptide sequencer from Applied Biosystems.
2.7. LC-MS/MS Analysis
The N-glycosylation site was analyzed using trypsin digestion followed by the addition of PNGase F (N-glycosidase F). Both deglycosylated and non-deglycosylated samples were subjected to LC-MS/MS analysis. LC-MS/MS was performed with Q Exactive using a Dionex Ultimate 3000 RSLC pump (Thermo Scientific, USA) coupled with an Acclaim PepMap RSLC C18 column (2 μm, 75 μm x 150 mm, Thermo Scientific, USA) at a flow rate of 0.25 μL/min using the gradient shown in Table S1. Xcalibur 2.2 was used to obtain the BPI chromatograms. Raw data were processed into peak lists by Thermo Proteome Discoverer 1.4 and searched in the Mascot database (Fig. S1A- S1C).
2.8. Periodic Acid-Schiff (PAS) Staining and VVL/PNA Staining Assays
Periodic acid-Schiff (PAS) staining for glycoprotein detection was performed using the Thermo Scientific Pierce Glycoprotein Staining kit (Thermo Scientific, USA). In the assay, protein samples were first fractionated on a 10% SDS-PAGE gel and then either stained with Coomassie blue for the control or treated with a staining kit according to the manufacturer’s instructions.
Biotinylated Vicia Villosa Lectin (VVL) and Peanut Agglutinin (PNA) purchased from Vector Labs (Vector Labs, USA) were used for Tn-antigen or T-antigen (mucin-type O-linked glycoprotein) detection, respectively. For detection, the protein samples were fractionated on gels and transferred onto nitrocellulose membranes. Then, to block nonspecific binding, the membranes were incubated in a carbo-free blocking solution (SP-5040) for 30 minutes then incubated in PBS containing 2-20 µg/ml biotinylated VVL or PNA for 30 minutes at room temperature. VECTASTAIN ABC-AP (alkaline phosphatase, AK-5000) reagents and BCIP/NBT (SK-5400) were applied according to the manufacturer’s instructions, resulting in black bands with a colorless background.
2.9. Ferrous Iron Chelating Capacity Assay
The ferrous ion chelating activity of the rice-expressed rpLF was investigated according to the method of Dinis et al. [50]. Briefly, purified rice-expressed rpLF was added to a solution of 2 mM FeCl2 and methanol. Then, the reaction was initiated by the addition of 5 mM ferrozine; the mixture was shaken vigorously then left standing at room temperature for 10 min. After the mixture had reached equilibrium, the absorbance of the solution was measured spectrophotometrically at 562 nm, where the Fe2+ chelating ability of rpLF was monitored by measuring the ferrous ion-ferrozine complex at 562 nm. The percentage of inhibition of ferrozine-Fe2+ complex formation was given by the following formula:
Ferrous ions chelating (%) = [(A0-A1)/A0] x 100
where A0 is the absorbance of the control, and A1 is the absorbance in the presence of the rpLF sample. The control contains FeCl2 and ferrozine complex formation molecules. In addition, EDTA was used as a positive control, and bLF was included for comparison.
2.10. Lipid Peroxidation Inhibition Assay
Lecithin (300 mg) was sonicated in an ultrasonic cleaner in 30 mL of 50 mM phosphate buffer (pH 7.4) for 4 hours. FeCl3 (0.5 mL, 1 mM), ascorbic acid (0.5 mL, 400 mM) and rpLF were then added to the sonicated solution (0.5 mL, 10 mg lecithin/mL). The mixture was incubated for 1 hour at 37°C, and oxidation was measured by the thiobarbituric acid (TBA) method [55]. The absorbance of the sample was read at 532 nm against a negative control, which contained all reagents except lecithin. The percent inhibition formula is given below:
Inhibition of lipid peroxidation (%)= [1- (A1/A0)] x 100
where A0 is the absorbance of the negative control reaction and A1 is the absorbance in the presence of rpLF. In addition, the synthetic antioxidant butylated hydroxytoluene (BHT) was used as a positive control, and bLF was included for comparison.
2.11. Reducing Power Assay
The reductive capability of pure rpLF was quantified by the method of Oyaizu [56]. The pure rpLF was mixed with phosphate buffer (0.2 M, pH 6.6) and potassium ferricyanide (1%). The mixture was then incubated at 50°C for 20 minutes, and the reaction was terminated by adding a portion of trichloroacetic acid (10%). The upper layer of solution was mixed with FeCl3 (0.1%), and the absorbance was measured at 700 nm in a spectrophotometer against ascorbic acid (Vit C), which was used as a control. The bLF was included for comparison.
2.12. DPPH (1,1-diphenyl-2-picrylhydrazyl) Scavenging Capacity Assay
The free radical scavenging activity of pure rpLF was measured by DPPH using the method of Blois [57]. A 0.1 mM solution of DPPH in ethanol was prepared, and pure rpLF at different concentrations was added. After 30 minutes, the absorbance of the reaction mixture was measured at 517 nm against BHT, which was used as a control. A low absorbance of the reaction mixture indicated high free radical scavenging activity and was calculated using the following equation:
DPPH scavenging effects (%)=100- [(A0-A1/A0) x100]
where A0 is the absorbance of the control reaction, and A1 is the absorbance in the presence of pure rpLF. The synthetic antioxidant butylated hydroxytoluene (BHT) was used as a positive control, and bLF was included for comparison.
2.13. Statistical Analysis
The results were evaluated as mean values of three times of the same sample, and data are represented as the mean ± standard deviation (mean ± SD).
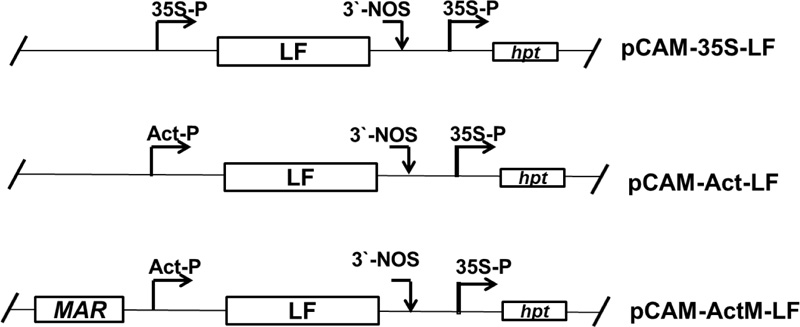
A) The relative amount of rpLF from the transgenic rice lines pCAM-Act-LF and pCAM-ActM-LF was compared to that of pCAM-35S-LF (set as 1). Data were measured from the Western blot results shown below.
B) Western blot analysis of rpLF from the transgenic rice lines pCAM-35S-LF, pCAM-Act-LF and pCAM-ActM-LF and non-transgenic (NT) rice. The same amount of total extractable protein (45 μg) was analyzed.
3. RESULTS
3.1. Analysis of rpLF Expression in Transgenic Rice
In the present study, the expression levels of recombinant porcine lactoferrin (rpLF) in transgenic rice lines driven by either the 35S promoter (35S-LF), actin promoter (Act-LF) or actin promoter in combination with tobacco nuclear matrix attach region RB7 MAR [58] sequences (ActM-LF) were evaluated (Fig. 1).
Through agrobacterium-mediated transformation, more than 20 independent transgenic rice lines were obtained for each construct, and at least 3 homozygous lines of each construct were selected for characterization for more than 10 generations. Typically, the homozygous lines with one T-DNA insertion event can be obtained within 2 or 3 generations of hygromycin selection, of which selection can be confirmed when approximately 20-40 progeny seeds from each generation are all germinated normally under hygromycin selection. The representative Western blot results of transgenic lines with three different constructs and their relative amounts of rpLF were measured and are presented (Fig. 2A, B). The expression levels of rpLF in ActM-LF transgenic rice were approximately 2 and 6 times higher than those in Act-LF and 35S-LF, respectively (Fig. 2B). Thus, the ActM-LF transgenic line was chosen for rpLF protein purification and further characterization.
3.2. Purification of rpLF from Transgenic Rice Grains
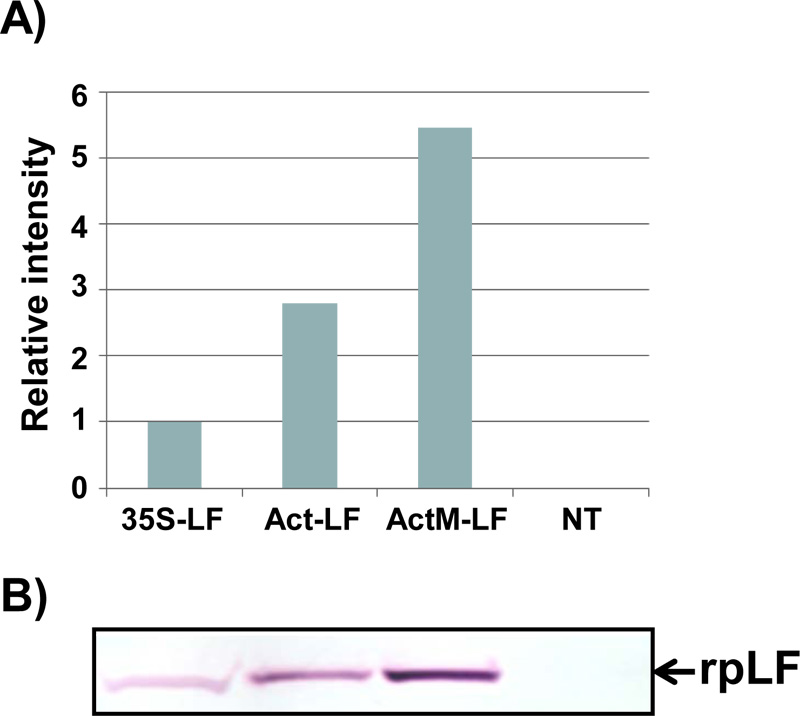
Purification of rpLF from rice grains yielded 815 mg of total extractable protein and 987 μg of rpLF, representing approximately 0.12% purity in this protein fraction (Table 1). These extracted proteins were further purified through 4-55% ammonium sulfate precipitation, and the rpLF purity reached 0.78% (6.5-fold improvement) with an approximately 25% loss of rpLF (Table 1). After rpLF was passed through the heparin-Sepharose column with an AKTA purifier, 326 μg of protein was eluted in the 0.34 M NaCl fraction, indicating approximately 100% purity and representing 33% recovery (Table 1). The purity of rpLF in all purification steps was analyzed by Coomassie blue-stained SDS-PAGE and Western blot assays (Fig. 3). In the Coomassie blue-stained gel, many protein signals appeared in the first three purification steps with 20 μg protein samples (Fig. 3A, lanes 1-3), and only one rpLF protein band was observed in the column-purified sample loaded with 1 μg of protein (Fig. 3A, lane 4). The column-purified rpLF was collected from eluted fractions which contains a higher concentration of the target protein, e.g. the fractions E4 to E9 (Fig. S3). These high concentrated protein fractions were then pooled and desalted and concentrated further through Amicon 30K UFC903008 (Merck Millipore, USA). In the Western blot assay, rpLF bands were observed for all protein samples loaded with either 20 μg for the first three steps (Fig. 3B, lanes 1-3) or 0.01 μg for the column-purified step (Fig. 3B, lane 4). In summary, column-purified proteins had an rpLF purity of approximately 100% based on the purification data and protein gel analysis results.
3.3. The Signal Peptide of rpLF Could be Correctly Processed in Rice
To determine if the rice-expressed rpLF could be correctly processed as it is in porcine, the purified rpLF was analyzed by the Edman degradation assay. The results of the Edman degradation assay revealed that the N-terminal sequence of rpLF starts with APKK (Fig. S1-A), demonstrating that the 19 amino acid signal peptide of rpLF could be removed correctly in transgenic rice.
3.4. rpLF was Glycosylated at Predicted N-glycosylation Sites, and Isoforms were Revealed in Rice
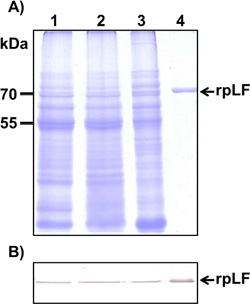
The same amounts of purified rpLF, horseradish peroxidase (a glycoprotein, used as a positive control), soybean trypsin inhibitor (negative control) and bovine lactoferrin (bLF) were fractionated on a 10% SDS-PAGE gel and stained with Coomassie blue (Fig. 4A) or treated with PAS (Fig. 4B). The results showed that rpLF is a glycoprotein (Fig. 4B, lane 3) as it was stained with PAS; however, rpLF contained fewer polysaccharides than bovine lactoferrin (Fig. 4B, compare to lane 4). Furthermore, the MS/MS assay demonstrated that two potential N-glycosylation sites located at N365 and N472 were glycosylated in the purified rpLF (Fig. S1B-S1C).
| Steps | Total protein (mg)b |
rpLF (µg)c |
Recovery (%) |
Purity (%)d |
| 1. Crude extract | 815 | 987 | 100 | 0.12 |
| 2. Supernatant of 4% ammonium sulfate | 734 | 972 | 98.5 | 0.13 |
| 3. Precipitate of 55% ammonium sulfate | 97 | 754 | 76.4 | 0.78 |
| 4. Heparin-Sepharose column | 326x10-3 | 326 | 33.0 | 100 |
b The concentration of protein was determined by the Bradford method
c The concentration of protein was determined by ELISA
d The purity of rpLF was measured by dividing c over b
A) Coomassie blue-stained SDS–PAGE for crude extraction (20 μg, lane 1), supernatants of 4% ammonium sulfate treatment (20 μg, lane 2), precipitates of 55% ammonium sulfate treatment (20 μg, lane 3), and column-purified rpLF (1 μg, lane 4). The molecular weight markers are indicated in kDa.
B) Western blot analysis of rpLF with the same amount of protein (20 μg) as above, except 0.01 μg of column-purified rpLF (lane 4) was used.
To determine the accurate molecular mass of rpLF and to check if rpLF contains multiple forms in transgenic rice, the MALDI-TOF MS assay was performed. At least two major peaks with molecular masses of 76652.5 and 77269.1, slightly heavier than the calculated molecular weight of 75350, were revealed in the assay (Fig. 5A). Similarly, the SDS–PAGE analysis resolved multiple bands for purified rpLF under nonreducing conditions (Fig. 5B), indicating the heterogeneity of the molecular mass of rpLF and corroborating the multiple-peaks observed by the MS assay.
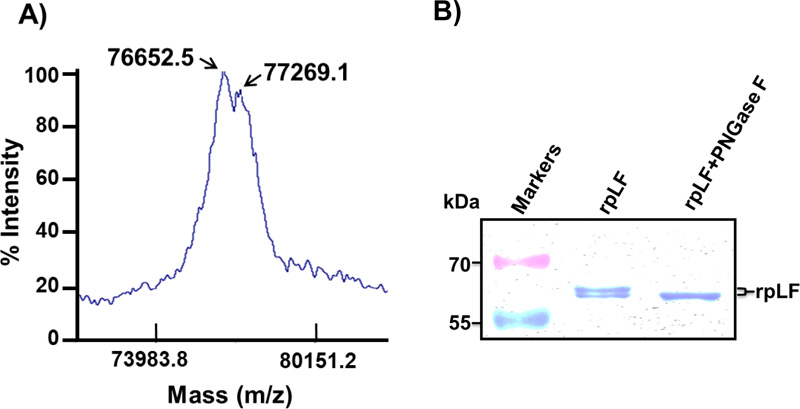
A) Purified rpLF was subjected to MALDI-TOF, and two peaks with molecular masses of 76.7 and 77.3 kDa were identified.
B) Coomassie blue-stained SDS–PAGE of purified rpLF (1 μg) and rpLF, which were incubated with PNGase F at 37°C for 2 hours (rpLF+PNGase F). The molecular weight markers are indicated in kDa.
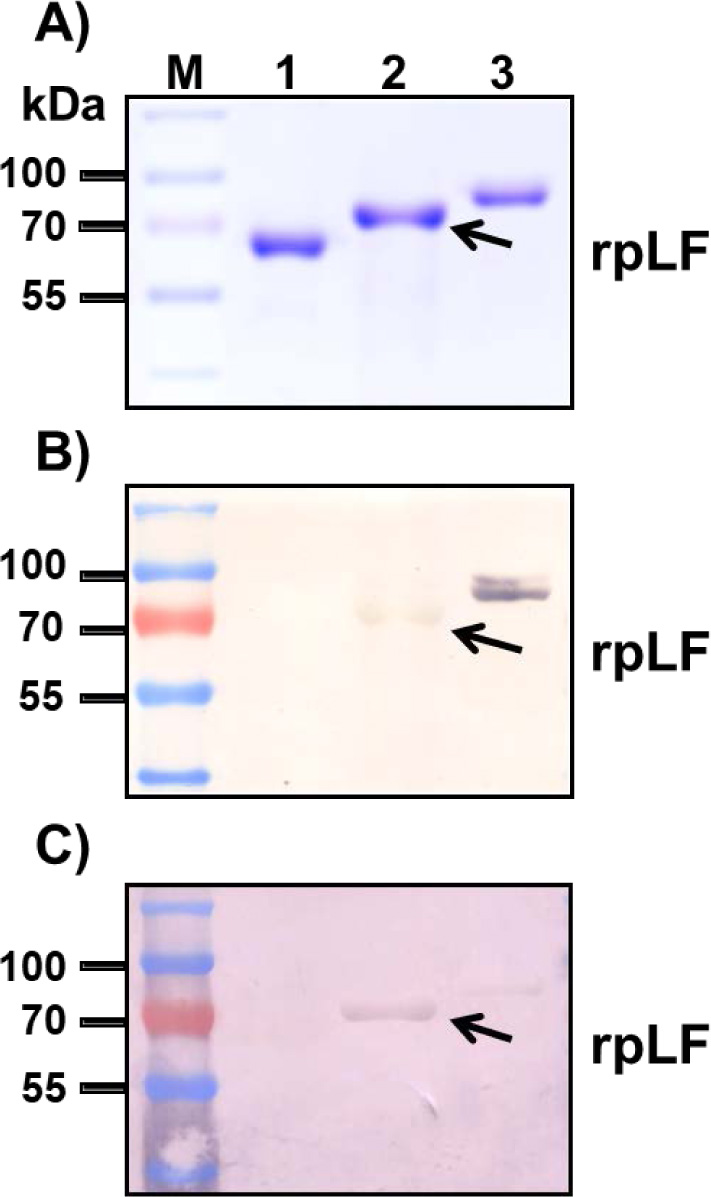
A) Polyacrylamide gel stained with Coomassie blue; the same amount BSA (lane 1), rpLF (lane 2, indicated by arrow) and bLF (lane 3) was subjected to SDS-PAGE and stained with Coomassie blue. Lane M, molecular mass markers.
B) Stained with VVL; the same protein samples stained with VVL. The arrow indicates rpLF.
C) Stained with PNA; the same protein samples stained with PNA. The arrow indicates rpLF.
To characterize whether these molecular mass isoforms were caused by glycosylation, rpLF was treated with PNGase F glycosidase and subjected to SDS–PAGE analysis. The results showed that the PNGase F glycosidase-treated rpLF formed a single band with a reduced molecular mass (Fig. 5B), indicating that glycans in rpLF were removed by PNGase and revealing different degrees of glycosylation.
3.5. Rice-produced rpLF Contains Mucin-type O-linked Glycans
To confirm that rpLF contains mucin-type O-linked glycans, biotinylated Vicia Villosa Lectin (VVL), which can detect the Tn antigen (GalNAcα-Ser/Thr), and Peanut Agglutinin (PNA), which can detect the T antigen (Galβ1-3GalNAcα1-Ser/Thr), were used as both the Tn antigen and T antigen are mucin-type O-linked glycoproteins. In these assays, both rpLF and bLF showed positive signals in VVL and PNA stains (Fig. 6B and C). rpLF showed a weaker signal than bLF in the VVL stain (Fig. 6B) but a stronger signal than bLF in the PNA stain (Fig. 6C), suggesting that O-linked glycosylation is present in rpLF and bLF and that more T-antigen glycosylation appeared in rice-produced rpLF.
3.6. Rice-produced rpLF Exhibited Antioxidant Activities
To confirm the antioxidant activity of rice-produced rpLF, four methods including 1) metal chelating capacity assay, 2) lipid peroxidation inhibition assay, 3) reducing power assay and 4) free radical scavenging capacity assay testing various molecular mechanisms were used.
3.6.1. Metal (ferrous) Ion (Fe2+) Chelating Capacity Assay
In this assay, EDTA and bLF were included as controls to compare the relative antioxidant activity of rpLF. The ferrous iron chelating ability of rpLF was approximately 63% at a concentration of 0.0625 mg/mL, increased to approximately 80% at a concentration of 0.25 mg/mL, and leveled off until 1 mg/mL (Fig. 7A), indicating that a saturation state was almost attained with 0.25 mg/mL rpLF. The chelating ability of EDTA was approximately 92% at a concentration of 0.0625 mg/mL, then leveled off until 1 mg/mL (Fig. 7A), indicating that a saturation state could be reached at 0.0625 mg/mL EDTA. In this assay, the chelating capacity of rpLF was 68 to 87% the chelating capacity of the synthetic metal chelator EDTA when 0.0625 and 0.25 mg/mL rpLF were compared. On the other hand, when the same concentrations were compared, the chelating ability of rpLF was higher than that of bLF. In summary, with the same low concentration of 0.25 mg/mL, the Fe2+-chelating ability of rice-produced rpLF was 87% that of EDTA and greater than that of bLF (Fig. 7A).
3.6.2. Lipid Peroxidation Inhibition Assay
In this assay, the activity of rpLF at different concentrations was investigated and compared to the activities of the synthetic antioxidant butylated hydroxytoluene (BHT) and bLF. The inhibition of lipid peroxidation by rpLF was approximately 19.8% at a concentration of 0.25 mg/mL and then increased to approximately 29.9% at a concentration of 1.0 mg/mL rpLF (Fig. 7B). The synthetic antioxidant BHT showed inhibition of 32.2% at a concentration of 0.25 mg/mL and an inhibition of approximately 84.9% at a concentration of 1.0 mg/mL (Fig. 7B). In comparison, the peroxidation inhibition activity of rpLF was approximately 35% that of BHT, similar to the inhibition activity of bLF.
3.6.3. Reducing Power Assay
In this assay, VitC and bLF were used as controls. The reducing power of rpLF measured at an absorbance of 700 nm (A700) was approximately 0.17 at a concentration of 1 mg/mL and then increased to approximately 0.70 at a concentration of 20 mg/mL (Fig. 7C). The reducing power of the VitC control was high (A700 = 2.38) at a concentration of 1 mg/mL, then increased gradually, but not proportionally, to approximately 2.88 at a concentration of 20 mg/mL (Fig. 7C). In comparison, the reducing power of rpLF was approximately 7 to 24%, similar to that of VitC. However, rpLF showed better reducing power activity than bLF (0.70 vs 0.47) at a concentration of 20 mg/mL (Fig. 7C).
3.6.4. 1,1-Diphenyl-2-picrylhydrazyl (DPPH) Free Radical Scavenging Capacity
In this assay, BHT and bLF were used as controls. The DPPH radical scavenging capacity of rpLF was approximately 39% at a concentration of 0.25 mg/mL and increased to approximately 46% at a concentration of 1 mg/mL (Fig. 7D). The DPPH scavenging capacity of BHT was approximately 94% at a concentration of 0.0625 mg/mL, then leveled off until 1 mg/mL (Fig. 7D), indicating that a saturation state could be reached as low as 0.0625 mg/mL. In this assay, the DPPH scavenging capacity of rpLF was approximately 41 to 49%, similar to that of BHT, at concentrations of 0.25 and 1 mg/mL. When the same concentrations were compared, the DPPH scavenging capacity of rpLF was slightly higher than that of bLF.
4. DISCUSSION
4.1. The Expression of rpLF in Transgenic Rice was Higher with the Rice Actin Promoter than the 35S Promoter
The present study compared the expression levels of rpLF in Act-LF to 35S-LF and ActM-LF, two additional transgenic lines. The expression level of rpLF in 35S-LF was approximately one-third (1/3) that in Act-LF. Although the CaMV 35S promoter has been widely used and well characterized in transgenic plants [59, 60], including dicotyledonous plants, such as tobacco [61], and monocotyledonous plants, such as rice [62], the levels of gene expression produced by this promoter in cereals were much lower than those in dicots [63]. In contrast, the rice actin promoter was an efficient promoter in transgenic rice [52] and other cereal crops [64]. The present study showed that the expression of rpLF in transgenic rice was higher with the rice actin promoter than the 35S promoter, which is consistent with previous reports [35, 63].
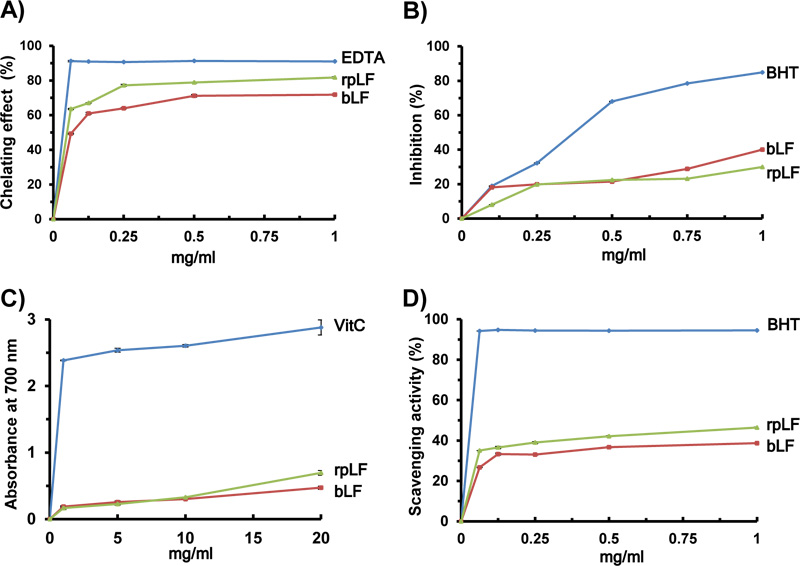
A) The ferrous iron chelating capacity of rpLF, bLF and EDTA.
B) Lipid peroxidation assay of rpLF, bLF and BHT.
C) Reducing power assay of rpLF, bLF and ascorbic acid (Vit C).
D) DPPH scavenging activity assay of rpLF, bLF and BHT. Each value represents the mean±SD (n=3).
4.2. Tobacco RB7 MAR Enhanced rpLF Expression in Transgenic Rice
Transgenes flanked with MAR sequences have been shown to be able to achieve a relatively higher level of transgene gene expression in transgenic plants [36-40]. Expression levels have been shown to increase up to 60 times using RB7 MAR in transgenic tobacco plants [36]. However, the same RB7 MAR sequences only resulted in an approximately 2.5-fold increase when applied in transgenic rice plants [40]. In the present study, the expression levels of rpLF in the transgenic rice line ActM-LF (with a vector driven by an actin promoter fused with one copy of the tobacco RB7 MAR DNA fragment) showed an approximately 2-fold increase compared to Act-LF, indicating that the effect of RB7 MAR on the expression of the porcine lactoferrin gene and the GUS gene in transgenic rice was similar, as previously observed [40].
4.3. rpLF at Nearly 100% Purity can be Obtained through Heparin-Sepharose Chromatography
While the transformation technology for producing recombinant pLF in transgenic rice has been well researched [31], the process to isolate pure rpLF for functional characterization and a robust manufacturing method have not been fully developed. In the present study, high salt crude extraction, ammonium sulfate cutting and column purification were utilized to achieve high levels of rpLF purification. The crude extraction with 20 mM Tris and 500 mM NaCl yielded total extractable protein at approximately 1% of the grain weight, which was less than the theoretical ~7% of total protein content, and the amount of rpLF determined by ELISA was approximately 0.12% of the total extractable proteins, which was 8 times less than the amount (approximately 1%) reported previously [31]. The low yield of rpLF and total extractable protein in the present study could be attributed to using a larger sample scale and different extraction buffer than in previous studies [31]. After crude extraction, purification was performed with 4-55% ammonium sulfate cutting, a series of empirical results, and a heparin-Sepharose column to reach nearly 100% purity with approximately 33% recovery from the crude extraction step. The chromatography step in this purification scheme removed most nontarget proteins and enhanced the purity of rpLF from 0.78% to 100% at the cost of approximately 55% loss in rpLF. In summary, the chromatography step significantly enhanced the purity of rpLF and shortened the purification period. Based on this purification scheme, a standardized purification protocol could be further developed as a basis for potential commercial production.
4.4. rpLF was Processed Post-translationally, and N-glycosylation Occurred at Different Degrees in Transgenic Rice
The present study demonstrated that the signal peptide of rpLF could be precisely removed and that rpLF is a glycoprotein in transgenic rice. The MS/MS assay further showed that two potential N-glycosylation sites located at N365 and N472 were glycosylated in the purified rpLF. Similarly, N-linked glycosylation and the glycan structures of a rice-derived recombinant human lactoferrin (rhLF) have also been characterized [18, 65]. Although the core glycan structure of rhLF is different from natural hLF, their biochemical and biophysical properties are similar [18]. In addition, rice-derived rhLF retains many of its biological activities [32]. A previous study also demonstrated that rice-derived rpLF retained iron-binding and antimicrobial activities [31]. All these observations support the notions that the post-translation process and N-glycosylation of recombinant rLFs, regardless of their origins, occurred in transgenic rice and that the biological activities of natural LF could be retained in rLFs.
In the present study, purified rpLF showed two different molecular mass peaks in the MALDI-TOF-MS assay and multiple bands in SDS–PAGE under nonreducing conditions, indicating the heterogeneity of the molecular mass of rpLF. These molecular mass differences were possibly caused by various degrees of post-translational modifications, such as glycosylation in transgenic rice. After treatment with PNGase F glycosidase, a single band with a reduced molecular mass was observed, suggesting that different degrees of rpLF glycosylation existed in transgenic rice. Taken together, the results indicate that the signal peptide of rice-produced rpLF could be correctly processed and that different degrees of N-glycosylated rpLF existed in the transgenic rice, causing molecular mass heterogeneity.
4.5. The Mucin-type O-glycosylation in Rice-produced rpLF requires Further Investigation
Although mucin-type O-glycosylation has not been described in plants [66], a report showed that protein subpopulations from alcohol-extracted rice grains were recognized by biotinylated VVL and PNA, suggesting that mucin-type O-glycosylation was present in rice [67]. A recombinant human GM-CSF derived from suspension-cultured rice cells showed various O-glycans attached to three O-glycosylation sites [68]. In the present study, the observations of rpLF with VVL and PNA blotting suggested that mucin-type O-glycosylation was present in rice and could imply that rice grains have the machinery to perform GalNAc O-glycosylation. However, since the detection of O-linked glycoprotein by VVL and PNA staining methods was indirect, the observations could be possible artifacts. Confirming whether rpLF contains mucin-type O-glycans and whether rice grain possesses machinery to perform GalNAc O-glycosylation will require further investigation.
4.6. The Antioxidant Activity of Rice-produced rpLF was Equivalent to that of Bovine LF
Although LFs from different animal sources, such as humans [48], bovines [45-47] and camels [49], displayed antioxidant activity, whether recombinant LF produced from transgenic plants also retains antioxidant activity has not yet been fully demonstrated. In the present study, the Trolox equivalent antioxidant capacity, DPPH free radical scavenging capacity, ferrous iron chelating capacity and reducing the power of rpLF were analyzed. The reducing DPPH radical-scavenging activity of purified rpLF was compared to that of bLF and BHT. rpLF showed the same radical-scavenging activity as bLF while possessing approximately 49% of the antioxidant properties of BHT. Further experimentation also showed that rpLF had a nearly equivalent capacity to bLF in terms of reducing power, iron chelating activity and lipid peroxidase activity.
LIST OF ABBREVIATIONS
| BHT | = Butylated Hydroxytoluene |
| bLF | = Bovine Lactoferrin |
| DPPH | = 1,1-Diphenyl-2-picrylhydrazyl |
| LC-MS/MS | = Liquid Chromatography-tandem Mass Spectrometry |
| LF | = Lactoferrin |
| MALDI-TOF | = Matrix-assisted Laser Desorption Ionization-time of Flight |
| MAR | = Matrix Attachment Region |
| NT | = Nontransgenic |
| PAS | = Periodic Acid-Schiff |
| PNA | = Peanut Agglutinin |
| VVL | = Biotinylated Vicia Villosa Lectin |
| rhLF | = Recombinant Human Lactoferrin |
| rpLF | = Recombinant Porcine Lactoferrin |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used in this study.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
SUPPLEMENTARY MATERIALS
Supplementary material is available on the publisher’s website along with the published article.
FUNDING
This work was financially supported in part by the Advanced Plant Biotechnology Center via the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the (MOE) (MOE-108S0021G and MOE-109S0021G) in Taiwan. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We thank the Division of Biotechnology, TARI, for providing the isolated field to grow the transgenic rice plants.


