All published articles of this journal are available on ScienceDirect.
Molecular Characterization of Fusarium Solani Degrades a Mixture of Low and High Molecular Weight Polycyclic Aromatic Hydrocarbons
Abstract
Objectives:
This study evaluates the ability of a non-white rot fungus strain, HESHAM-1, to degrade a mixture of low (naphthalene and phenanthrene) and high (chrysene and benzo(a)pyrene) molecular weight polycyclic aromatic hydrocarbons (LMW and HMW PAHs).
Methods:
Strain HESHAM-1 was isolated from oil polluted soil by enrichment method using phenanthrene as the sole source of carbon and energy. The strain showed the ability to tolerate and degrade a mixture of both low and high molecular weight PAHs. In the presences of LMW-PAHs (naphthalene and phenanthrene) as co-substrate, chrysene and benzo(a)pyrene (HMW-PAHs) were, respectively degraded by the fungus strain HESHAM-1 which was confirmed by GC-MS analyses.
Results:
The degradation rate was found as 84.82% for naphthalene, 40.09% for phenanthrene, 57.84% for chrysene and 71.06% for benzo(a)pyrene at the end of 10 days. This is the first report describing the biodegradation of a mixture of four PAH compounds by non-white rot fungus strain HESHAM-1 isolated from Egyptian oil-polluted soil. The fungus strain HESHAM-1 was identified by morphological characteristics and molecular genetics technique based on PCR amplification and sequencing of the internal transcribed spacers (ITSs) of the rDNA region and intervening 5.8S rRNA gene. Blast result and phylogenetic analysis of gene sequencing suggested that strain HESHAM-1 was closely related to Fusarium solani with 100% sequence identity.
Conclusion:
The present study clearly demonstrates that, strain HESHAM-1 could be used to remove the crude oil from the environment.
1. INTRODUCTION
Polycyclic Aromatic Hydrocarbons (PAHs) are a group of complex environmental pollutants that are composed of carbon and hydrogen with fused benzene rings in linear, angular, and clustered arrangements. These compounds were classified according to their molecular weight as low molecular weight PAHs (LMW PAHs), which consist of two or three rings such as naphthalene and phenanthrene, and high molecular weight PAHs (HMW PAHs), which consist of the more than three aromatic rings like pyrene, chrysene and benzo(a)pyrene. PAHs are widespread throughout environment, being found in the air, water and soil. They can also be found in various substances such as crude oil, coal, coal tar pitch, creosote, roofing tar and anthracene oil, which are commonly used pesticides for wood treatment[1-4]. PAHs and their metabolites are documented to have mutagenic and carcinogenic characteristics for humans and animals [5, 6]. Therefore, the removal of PAHs from the environment is a major concern since they can exert acute as well as chronic toxicity [7].
HMW-PAHs are less water soluble and more recalcitrant to degradation than LMW-PAHs [8]. Many strategies were used to remove PAHs, such as physical treatment (adsorption, volatilization and photolysis), chemicals [9-12] and microbial degradation using bacteria, fungi, algae and yeasts. Microbial degradation is more preferable due to cost effectiveness and complete mineralization of PAHs to safe products [13]. Various microorganisms including fungi, algae, yeasts and bacteria can degrade PAH compounds [14-17].
Fungi are tolerant to high concentrations of recalcitrant compounds and are able to flourish in extreme conditions, such as at high temperatures and under low pH conditions. In addition, the fact that fungi are capable to grow and distribute through a solid matrix to degrade PAHs within contaminated areas may be due to its ability to form large, branching mycelia [18, 19]. Several fungi are known to have the ability to degrade persistent pollutants [20]. Most of the previous studies on biodegradation of PAHs by fungi have focused on white-rot fungi [21-24]. Boyle et al. [25] found that white-rot fungi growing in soil could not breakdown significant amounts of PAHs. However, there is limited knowledge about the degradation of PAHs by non-white rot fungi, the aim of this study was to isolate and examine the non-white-rot fungi that could degrade a mixture of four compounds representatives of LMW and HMW-PAHs. Sequence analysis of the 5.8S rRNA gene and the internal transcribed spacer regions were used for fungus identification.
2. MATERIALS AND METHOD
2.1. Chemicals and Samples
All PAHs, including LMW (naphthalene, phenanthrene) and HMW (chrysene, benzo(a)pyrene) were obtained with high purity from Acros Organics Co. (Geel, Belgium). Four Soil samples contaminated with oil were collected from various petroleum station sites in Assiut governorate, Egypt.
2.2. Enrichment and Isolation of PAH-Degrading Fungi
Soil enrichment technique using mineral basal salts (MBS containing, per liter of distilled water, 1000 mg (NH4)2SO4, 800 mg K2HPO4, 200 mg KH2PO4, 200 mg MgSO4·7H2O, 100 mg CaCl2·2H2O, 5 mg FeSO4·7H2O, pH 5-6, and 1 mL of vitamin solution sterilized by filtration) medium supplemented with 0.2% phenanthrene was used as a carbon source for the isolation of PAH-degrading fungi, as described previusly by Hesham et al. [14]. At the end of the enrichment process, fungal strains were isolated using MBS agar plates coated with phenanthrene as the sole carbon source [14, 26].
2.3. Assessment of the Tolerance Ability of Isolated Fungi to PAHs
All fungal isolates were tested for their tolerance to selected PAHs by culturing onto MBS agar plates supplemented individually with 30 mg L−1 each of naphthalene, phenanthrene, chrysene and benzo(a)pyrene. As a control, isolates were inoculated on MBS agar plates supplemented with 2% glucose. The plates were incubated at 27 °C for a period of 7 days. The tolerance rate was estimated in triplicate using the following equation:
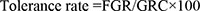 |
Where, FGR is the fungal growth rate in the presence of a certain PAH and GRC is the growth rate of a control culture. The FGR and GRC were calculated by measuring the diameters of the expanding colonies.
2.4. Molecular Identification of Fungal Isolate
2.4.1. Isolation of Genomic DNA and PCR Amplification of Fungal ITS Region
Genomic DNA was isolated from selected fungus by using DNeasy plant mini kit (Qiagen, USA) according to the company instructions. The purity and quantity of DNA was examined by recording its UV absorption spectrum and running a sample on a 1% agarose gel electrophoresis. For identification, the ITS region of fungal DNA was amplified using the fungal specific primer set: ITS1-F (5ʹ-CTTGGTCATTTAGAGGAAGTAA-3ʹ) and ITS4-R (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) as described by White et al. [27]. PCR was performed in a final volume of 50 μL containing GoTaq (Promega, Madison, WI, USA) green master mix, 1 µL genomic DNA sample and 1 µL each primer (at a concentration of 0.5 mM) [15]. The PCR conditions were as the following: initial denaturation at 95°C for 5 min, followed by 40 cycles at 94°C for 1 min, 55°C for 1 min, 72°C for 1 min; final extension at 72°C for 10 min and holding at 4°C.
2.4.2. PCR Products Purification and Sequence Determination
To verify the presence of appropriate sized amplicons, the PCR product for the isolate was subjected to electrophoresis in 1% agarose gel in TAE buffer according to the standard method. Product of the correct size was purified with a TakaRa Agarose Gel DNA Purification Kit Ver.2.0 and sequenced in both directions using an ABI 3730 automated sequencer (Macrogen, Seoul, Korea).
2.4.3. Alignment and Phylogenetic Analysis
The obtained sequence was aligned with known ITS sequences in GenBank database using the basic local alignment search tool (BLAST) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/), and the percent identity scores were generated to identify the fungus isolate. Phylogenetic tree was constructed with MEGA version 4.0 using a neighbor-joining algorithm, plus the Jukes-Cantor distance estimation method with bootstrap analyses for 1,000 replicates was performed [28].
2.4.4. Degradation of a Mixture of LMW and HMW-PAHs by Fungus Strain HESHAM-1
In a study by Andres, et al. [29], for preparation of PAH stock solutions, 100 mg from the each selected PAH (naphthalene, phenanthrene, chrysene benzo(a)pyrene) was dissolved in 10 mL acetone, added to 50 g Tween 80 in 50 mL distilled water, ultrasonicated for 10 min and then the suspension diluted with water to make 200 ml. Degradation experiment was performed using Erlenmeyer flasks (250 mL) containing 100 mL of MBS liquid medium. The fungus isolate that was selected according to the highest tolerance to PAHs was inoculated into the flasks containing MBS supplemented with the mixture of PAHs. The initial concentration of each PAH in the mixture was: naphthalene, 6.36 mg L–1; phenanthrene, 6.55 mg L–1; chrysene, 1.54 mg L–1; and benzo(a)pyrene, 4.61 mg L–1. Culture medium inoculated with autoclaved fungus and flasks containing uninoculated medium were used as negative and blank controls, respectively. Fungal cultures were grown in the dark with tumble-shaking at 150 rpm and 27°C [30]. The experiment was carried out in triplicates.
2.4.5. GC-MS Analysis
Hexane (10 mL) was added to each culture, shaken horizontally for 2 h at 150 rpm and approximately 2 mL of the hexane layer was placed in an autosampler vial, crimp-sealed with a Teflon-lined cap, and then stored at -20°C until analysis [31]. PAH concentrations were analyzed by gas chromatography interfaced with mass spectrometer detector (GC/MS), equipped with a 5973 mass selective detector under the selected ion-monitoring (SIM) mode [14]. The percentage of degradation was calculated by the following formula.
Degradation rate (%) = (Initial PAH concentration − Final PAH concentration)/Initial PAH concentration×100
2.4.6. Nucleotide Sequence Accession Number
The partial ITS/5.8S sequence of strain HESHAM-1 reported in this paper has been deposited in the DDBJ (www.ddbj.nig.ac.jp/), EMBL (www.embl.de/), and GenBank nucleotide sequence databases (http://www.ncbi.nlm.nih.gov) under accession number KP899543.
3. RESULTS AND DISCUSSION
3.1. Isolation of PAHs Degrading Fungi by Enrichment Technique
PAHs as hazardous compounds, that have varied effects on the environment, are recalcitrant and require specific microorganisms to perform the degradation for the contaminants or their transformation into less harmful non-hazardous compounds. Microorganisms offer an enormous field of environment friendly options for mineralization of contaminants or their transformation into less harmful non-hazardous compounds. Recently, researchers have been focusing on the degradation of HMW-PAH which has resulted in the isolation of a number of indigenous microorganisms that can mineralize and grow on four or five-ringed PAHs as the sole carbon and energy source [14, 15, 32, 33]. Enrichment technique is a way to obtain the promising indigenous microorganisms that could utilize PAHs as the sole carbon source [14]. In this study, 10 indigenous PAH degrading fungi were isolated from oil-polluted soils using enrichment culture technique. Fungal colonies were isolated on MBS agar plates coated by phenanthrene, as a sole source of carbon and energy. The level of tolerance of these fungal isolates to 2-5-ringed PAHs (30 mg L− 1) including, naphthalene, phenanthrene, chrysene and benzo(a)pyrene was investigated. Although all the fungal isolates were able to grow on MBS-agar plates coated by LMW-PAHs with high level of tolerance (data not shown), only one isolate designated as strain “HESHAM-1” exhibited the highest tolerance for the HMW-PAHs (benzo(a)pyrene) with the great growth on the plates. Therefore, “HESHAM-1” was selected as HMW-PAHs degrader.
Carrera [34] reported that the PAH tolerance test should be the primary factor in identifying desirable PAH-degrading fungal species for biotechnological procedures. Other studies demonstrated that the microorganisms, bacteria or yeasts, which isolated on naphthalene as the sole carbon source were unable to grow on anthrancen, phenanthrene, fluranthene, pyrene or chrysene, whereas microorganisms isolated on any of the other five compounds were capable of utilizing at least one of the other substrates [14, 35, 36]. On the other hand, the use of phenanthrene as a sole source of carbon has been chosen as a best way for selection of HMW-PAH degrading yeast strains [14].
3.2. Molecular Identification and Phylogenetic Relationship of the Fungus Strain HESHAM-1
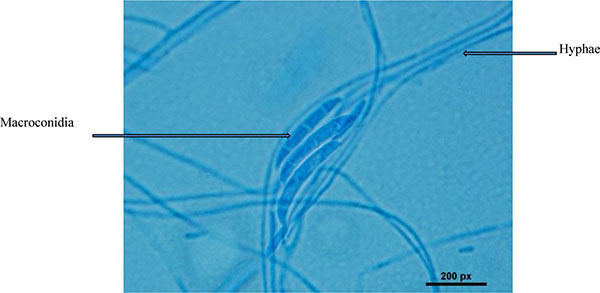
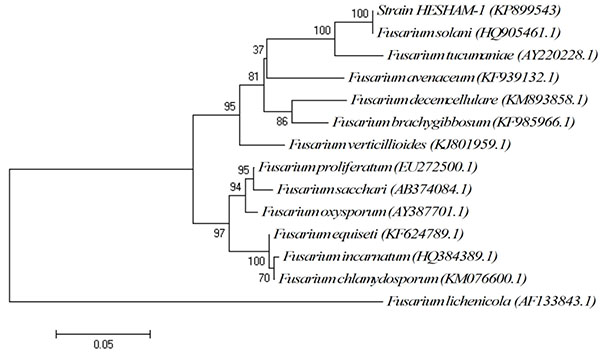
The conventional phenotypic method for identification of microbial species is often laborious and time-consuming and is hindered by the unstable and subjective nature of phenotypic characteristics, which are readily influenced by culture conditions. In contrast, the identification methods by molecular genetics techniques are rapid, easy and more precise for microorganism's identification [15, 16]. Strain HESHAM-1 was identified based on its morphological and microscopic examination as Fusarium sp. (Fig. 1). To confirm the identity of the fungus HESHAM-1 at the molecular level, the ITS1 and ITS4 primers were used to amplify the partial sequencing of ITS region by PCR. Fungal rDNA-ITS sequences for the HESHAM-1 strain were compared with the sequences of ITS region in GenBank database by means of BLAST search. Results show that the ITS sequence of the isolated strain was highly homologous to Fusarium solani, with 100% sequence identity (Fig. 2). To confirm the position of the strain HESHAM-1 in phylogeny, a number of sequences representative of some Fusarium sp. were selected from Genbank database for the construction of a phylogenetic tree. As shown in Fig. (3), the phylogenetic tree indicated that strain HESHAM-1 and Fusarium solani shared one clade cluster. Therefore, the strain HESHAM-1 was identified as F. solani. Recently, F. solani was identified based on ITS sequencing and phylogenetic analysis.

3.3. Biodegradation of LMW and HMW-PAHs by F. solani Strain HESHAM-1
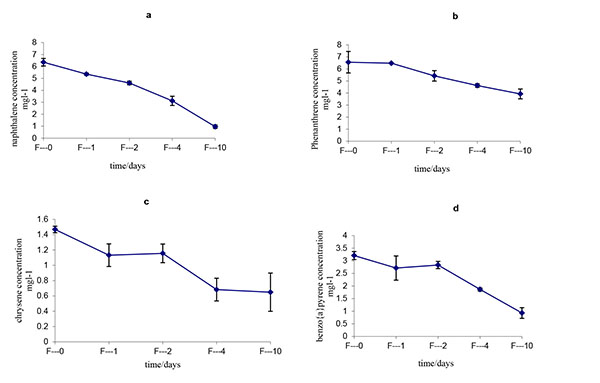
The initial and residual concentrations and degradation ratios of all of the LMW-PAHs and HMW-PAHs by Fusarium solani strain HESHAM-1 are described in Table 1 and Fig. (4). The data indicate that all of the PAHs in the mixture were degraded significantly by the strain HESHAM-1. The removal efficiency for the LMW-PAHs (naphthalene and phenanthrene) ranged from 40.09% – 84.82% and that for the HMW-PAHs (chrysene and benzo(a)pyrene) ranged from 57.84% – 71.06% after degradation for 10 days (Fig. 4). Of the microorganisms recognized to have the ability to degrade PAHs in the environment, fungi have been shown to be more successful than bacteria in breaking down the higher molecular weight compounds [37]. Fungi can degrade HMW- PAHs, whereas bacteria are best at degrading smaller molecules [23]. Most of the previous studies on biodegradation of PAHs by fungi have focused on white-rot fungi [21-24]. Boyle et al. [25] found that white-rot fungi growing in soil did not degrade significant amounts of PAHs. Fusarium solani, a non-white-rot species, is little known for its ability of HMW-PAHs degradation compared with white-rot fungi [38]. Researchers reported that Fusarium sp. isolated from PAHs-contaminated soil could degrade 250 mg L-1 as initial concentration of anthracene, phenanthrene and pyrene, when growing accompanied with five different bacterial species [38]. Hidayat and Tachibana [39] demonstrated that Fusarium sp. F092 have the ability to degrade chrysene and the aliphatic fraction of crude oil contaminating liquid culture with artificial sea water (35%). Rafin et al. [40] showed that the fungus F. solani is able to grow in liquid medium with benzo(a)pyrene as sole carbon source. Another report indicates that Fusarium sp. E033 demonstrated a greater biodegradation efficiency than that of F. solani, which degraded 6.8% benzo(a)pyrene over 15 days of incubation [41]. It is interesting that our results showed the HMW- PAHs including chrysene and benzo(a)pyrene as HMW-PAHs, were removed with a relatively high efficiency (57.84% – 71.06%) by F. solani strain HESHAM-1. It means that this fungus strain could able to degrade crude oil which consists of aliphatic and aromatic hydrocarbons and consumption of its components. The efficient removal of these compounds should be removed through co-metabolism by the fungus strain HESHAM-1 using the LMW-PAHs as the carbon sources. Hesham et al. [14] found that the 5 ring benzo(a)pyrene decomposed by Pichia anomala only through co-metabolism.
| PAHs | No. of Rings | Initial Concentration (mg/kg) | Remaining (mg/kg) | % Degradation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Naphthalene (LMW) | 2 | 6.355345 | 0.964706 | 84.82% | |||||||
| Phenanthrene (LMW) | 3 | 6.554856 | 3.926388 | 40.09% | |||||||
| Chrysene (HMW) | 4 | 1.538108 | 0.648375 | 57.84% | |||||||
| Benzo(a)pyrene (HMW) | 5 | 3.208544 | 0.928482 | 71.06% | |||||||
CONCLUSION
This is the first report describing the biodegradation of a mixture of four PAH compounds (naphthalene, phenanthrene, chrysene and benzo(a)pyrene) by a non-white rot fungus strain HESHAM-1 isolated from Egyptian oil-polluted soil. By sequencing of the 5.8S rRNA gene and the internal transcribed spacer regions, the strain HESHAM-1 was identified to be F. solani. The ability of this fungus strain to tolerate and degrade a mixture of LMW and HMW PAHs could recommend it for the biodegradation of crude oil-polluted sites.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
Decleared none.


