All published articles of this journal are available on ScienceDirect.
Isolation and Characterization of a Thermostable Cellulase from Bacillus licheniformis Strain Vic Isolated from Geothermal Wells in the Kenyan Rift Valley
Abstract
Cellulolytic microorganisms such as fungi and bacteria are responsible for much of the cellulose degradation in soils. Despite the vast number of cellulase producers, there is a deficiency of microorganisms that can produce significant amount of the three components of a cellulase system. i.e. CMCases, exoglucanases and β-glucosidases to efficiently degrade cellulose to fermentable products.
Soil samples were collected from two sites of the Kenyan Rift valley; Hell’s gate National Park and a geothermal well in Eburru hill at depths of 155 m, 156 m and 157 m. Isolation of cellulolytic bacteria was carried out using minimal media supplemented with 1% CMC. Isolates obtained were characterized using both morphological and molecular methods. A quantitative cellulolytic test was carried out to select the isolate with the highest specific activity. Additionally the cell-free supernatant was used for functional studies to determine the isolate’s cellulase system components, optimum temperature, pH and time stability.
Phylogenetic analyses indicated that all isolates associated mainly with members of the Bacillus licheniformis. All isolates were capable of utilizing CMC, avicel and Cellobiose as their sole carbon sources. Isolate 3 from a depth of 156 m in the well was identified as Bacillus licheniformis strain vic and selected for further functional studies. This isolate was found to bear the three enzyme components of a cellulase system. A cell free extract was noted to hydrolysecellobiose, Avicel and CMC with enzyme specific activities of 0.46878 U/mg, 0.18784 U/mg and 0.13571 U/mg respectively. Optimum temperature for activity measured over 60 minutes was found to be 60oC with relatively high activity at both 70oC and 80oC. The optimum pH at the predetermined optimum temperature was found to be pH 5. The crude enzyme extract was stable for eight hours of incubation at 60oC and pH 5 retaining 92.2% of its activity. This makes B.licheniformis strain vic a quite attractive isolate for potential application in industries.
INTRODUCTION
Cellulose is the most abundant plant biomass on earth with an estimated annual production of 4.0x107units [1]. This abundance confers to cellulose an immense potential as a renewable source of energy. Complete degradation of cellulose requires a suite of enzymes collectively referred to as cellulases. These are hydrolytic enzymes that hydrolyze the β -1,4-glucosidic linkages to smaller oligosaccharides and eventually glucose. This ability to hydrolyse β -1,4-glucosidic linkages distinguishes cellulases from other hydrolases. A cellulase enzyme system comprises of three classes of enzymes; CMCases (EC 3.2.1.4), exoglucanases (EC 3.2.1.91) and β-glucosidase (EC 3.2.1.21). This enzyme system acts in synergy to release glucose as the end product [2].
Cellulases are produced by a large diversity of microorganisms during their growth on cellulolytic substrates [3]. Some insects, crayfish and mollusks produce their own cellulases to utilize cellulose [4, 5]. Despite the vast number of cellulase producers, there is a deficiency of microorganisms that can produce significant amount of the three cellulase enzyme components i.e CMCase, exoglucanase, and β-glucosidase to efficiently degrade cellulose to fermentable products [6].
Cellulases have a wide range of application in numerous industries such as agriculture, pulp and paper, textile, food and beverage, animal feed, detergent industry and bioconversion of cellulosic material into solvents such as ethanol amongst others [7]. Tolerance to high temperatures is a highly desirable property for industrial cellulose degrading enzymes [8] since high temperatures are often required in these industries.
Culturable cellulolytic bacteria have been isolated from a wide array of environments ranging from soil, lakes [9], decomposing material [10], animal dung [11, 12], rumen of ruminants and termite gut [13-15], goldmines [16] and hot springs [17]. This clearly shows that bacteria have a wide range of natural habitats and this endows them with the capability of producing stable enzymes.
In this study traditional microbiological isolation techniques were used to screen for cellulose degrading bacteria. The isolates were identified and that bearing the highest enzymatic activity, selected for further characterization on its catalytic strategy on cellulosic material. Temperature and pH are amongst the factors that affect the stability of enzymes. Other factors include binding of metal ions, oxidative stress, solvents and presence of surfactants [18].
MATERIALS AND METHODS
Carboxymethylcellulose [CMC], avicel, cellobiose, soil samples
Isolation of Cellulose Degrading Bacteria
Soil samples from Eburru and Hell’s gate were separately suspended in physiological saline (0.85% NaCl) to suspend the cells. Aliquots of these preparations were inoculated in a selective medium containing 5g/l Carboxymethylcellulose (CMC), 2.2 g/LK2HPO4, 1.5g/L KH2PO4, 1.3 g/L (NH4)2SO4, 0.1 g/L MgCl2, 0.02 g/L CaCl2 and 0.001 g/L FeSO4. 7H2O [19]. Incubation was done at 45o C for 7 days. Further isolation was carried out by plating on CMC agar plates of selective medium (pH 6.9) supplemented with 0.1% yeast extract and 1.5% agar. Overnight incubation was done in a thermoshaker at 45oC and 150 rpm.
Screening for Cellulolytic Activity
Cellulolytic activity of the isolated bacteria was determined on a solid medium containing 5gL-1 CMC, 2gL-1 Yeast extract, 5gL-1 Malt extract and 15gL-1 Agar, pH 7.2 (modified fromDSMZ medium 65 without CaCO3 and glucose) [20] and incubated for 3 days at 45oC for secretion of cellulases. Cellulolytic activity was detected by soaking the plates with 1 mg/ml congo red for 15 minutes followed by destaining with 1M NaCl. Clear zones of hydrolysis were seen against the red color of Congo red stain indicating cellulose degradation.
Characterization of Strains
Morphological characterization of the strain was carried out via Gram stain and determination of colony characteristics such as shape, color, surface elevation and margins. Temperature and pH tolerance tests were carried out at ranges of 4oC to 60oC and pH 3-12, respectively. Growth on selective media supplemented with 5 gL-1 avicel and 5 gL-1 cellobiose was determined.
Analyses of 16S rRNA Gene Sequences
Overnight cultures of identified bacterial isolates, inoculated in nutrient broth were used for DNA extraction according to procedure described by Kate Wilson [21] was used to harvest total genomic DNA from the bacterial cells. About 100 ng of DNA from each isolate was used as a template for 16S rRNA gene amplification using two bacterial specific primers: 16sF27, forward 5’…AGA GTT TGA TC (AC) TGG CTC AG…3’ and 16sR 1492, reverse 5’…TAC GG (CT) TAC CTT GTT ACG ACT T…3’. The amplification was performed in 50μl reaction volume containing 36.5μl PCR grade water, 5μl 10× PCR buffer, 4μl MgCl2 (25mM), 1μl dNTP mix (10mM), 1μl of each primer (200μM), 0.5μl Taq polymerase (5U/μl) and 1μl of isolated DNA.
Polymerase chain reaction was performed in a TProfessionalthermocycler (Biometra, Germany). Conditions for the PCR were as follows; An initial denaturation step at 94ºC for 5 minutes followed by 35 cycles of 94ºC for 1 minute, 55ºC for 1 minute and 72ºC for 2 minutes and a final extension step of 7 minutes at 72ºC. The amplified products were electrophoresed on a 1% agarose gel in 1×TAE buffer containing 0.5 μg/ ml of ethidium bromide and visualized under UV light to determine the quality and size. The PCR products were then gel purified using MinElute Gel Extraction Kit (QIAGEN) according to manufacturer’s protocol and sent to International Livestock Research Institute (ILRI) Kenya for sequencing.
A consensus sequence for each isolate was generated using Chromas Lite software and the resulting sequences matched with known 16s rRNA gene sequences at NCBI using the BLAST algorithm.
Alignment of 16S rDNA Sequences
The 16S rDNA sequences obtained were aligned using Jalview 2.8 to show sequence variation. Additionally these DNA sequences were used to retrieve related sequences from gene bank for determination of inferred relationship. All the sequences were then aligned in MUSCLE [22] and a phylogenetic tree constructed based on the nucleotide sequences with the Bayesian phylogenetic method in MrBayes software [23]. The trees were then visualised using fig tree software obtained at http://tree.bio.ed.ac.uk/
ENZYME ASSAYS
Enzyme Production
For enzyme production, the four isolates were separately inoculated in 100 mL of nutrient broth media supplemented with 1% CMC Modified [24], and cultured for 12 hours at 45oC and 150 rpm. The culture was then centrifuged at 4000 rpm 4oC for ten minutes and the cell free supernatant collected as the crude enzyme for assays. Protein concentration (mg/ml) of the cell free supernatant was determined using the Biuret method and the values obtained used to calculate specific activity of the crude enzyme in respective enzyme assays.
Quantitative Carboxymethyl Cellulase Assay (CMCase) Assay
A quantitative Carboxymethyl cellulase (CMCase) assay for all four isolates was carried out using, 3, 5- Dinitrosalicylic acid (DNS) test [25]. A 1.0% (w/v) solution of carboxymethylcellulose (CMC) in 50mM Sodium acetate buffer (pH 5) was used as substrate. The reaction mixture (1 mL), containing 500μL crude enzyme and 500μL substrate, was incubated at 50oC for 10 min. The amount of reducing sugars produced was then determined usingDNS test with a glucose standard. Heat inactivated (100oC for 15 minutes) crude enzyme and substrate were used as controls.
Determination of Optimum Time for CMCase Production
Optimum time for CMCase production was determined by growing the isolate 3 in a minimum medium with 5gL-1 of CMC as the sole carbon source and incubation was carried out for 10 days at 45oC, pH 6.9 and 150 rpm. The Quantity of reducing sugars produced was assayed for ten days using the DNS test.
Determination of Isolate’s Cellulase System Components
Presence and level of exoglucanase and β-glucosidase activities in addition to CMCase activities for isolate 3 were determined. Assay preparations included 500μl of crude enzyme, and 1% Avicel, cellobiose and CMC prepared in 50mM Sodium acetate buffer pH 5 respectively and incubation carried for 30 minutes at 60oC. Quantity of reducing sugar (mg/ml) produced was determined using the DNS test.
Determination of Optimum Temperature
Effect of temperature on CMCase activity was determined from 30oC - 90oC at intervals of 10 oC. Reactions were prepared as follows; 1% CMC in 50mM sodium acetate buffer pH 5 and incubated in a water bath for 60 minutes for the selected temperatures. Heat inactivated crude enzyme and substrate were included. One enzyme unit (U) was defined as the amount of enzyme producing 1 μmol of reducing sugar measured as glucose per minute per mL.
Determination of Optimum pH
Effect of pH on enzyme activity was determined by mixing 500μL of the crude enzyme extract in each case with 500μl of 1% (W/V) CMC broth substrate, with pH values ranging from 3 -11.
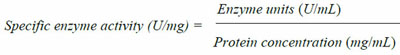 |
The buffers used for pH adjustment included Glycine-HCl (pH 3.0), HAc-NaAc (pH 4.0 - 5.0), Na2HPO4-NaH2PO4 (pH 6.0 - 8.0) and Glycine-NaOH (pH 9.0 - 11.0) buffers at a final concentration of 50mM (modified [24] the reaction mixture was incubated in a water bath at pre-determined optimum temperature of 60oC for 1 hour. Specific enzyme activity was then calculated.
RESULTS AND DISCUSSION
This study aimed at isolation of thermotolerant cellulose degrading bacteria from soil samples collected from Hell’s gate and a well in Eburru hills in the Kenya’s Rift valley. All four isolates obtained were gram-positive rods (Table 1). The 16S rDNA sequence analysis and phylogenetic tree construction revealed that the isolates clustered with members of Bacillus licheniformis (Fig. 2). Growth conditions studies showed that the isolates were able to grow in a wide range of pH and temperature conditions i.e. pH 3 - 12 and at 20 oC - 55oC, respectively. Optimum growth temperature range was determined as 40 oC - 50 oC with an optimum growth pH range of 5 - 6 (Table 1).
After isolation, a temperature of 45oC was selected for culturing bacterial isolates for subsequent studies. This temperature was used in a similar study for the isolation of cellulolytic bacteria among them being Bacillus licheniformis from bovine rumen [13] and Bacillus strain M-9 from decomposing rice bran straw [10]. Relatively high optimum growth temperatures for these isolates directly relates to their ability to produce enzymes that can tolerate high temperatures.
A qualitative test for presence of cellulolytic activity confirmed that all four isolates bear cellulolytic activity from the clearance zone observed around the colonies (Fig. 1). Additionally quantitative assays carried out revealed that isolate 3 (Bacillus licheniformis strain vic) had the highest activity (Fig. 3) as indicated by its ability to produce the highest reducing sugar content. Bacillus licheniformis strain Vic was subsequently selected for further studies.
Optimum time required for CMCase production is five days (Fig. 4) on minimum medium supplemented with CMC as the sole carbon source. Such a phenomenon implies that production of CMCase from Bacillus licheniformis strain vic using a minimum medium for industrial application would be time costly hence supplementation is necessary.
Colony characteristics, gram stain, pH and temperature growth ranges and substrate utilization for growth (avicel and cellobiose).
| Colony characteristics | Isolate 1 | Isolate 2 | Isolate 3 | Isolate 4 |
|---|---|---|---|---|
| Shape | Irregular | Irregular | Irregular | Irregular |
| Color | Cream white | Cream white | Cream white | Cream white |
| Elevation Surface | Raised Smooth | Raised Smooth | Raised Smooth | Raised Smooth |
| Margin | Lobate | Lobate | lobate | lobate |
| Gram stain Growth pH range |
+ ve rod |
+ ve rod |
+ ve rod |
+ ve rod |
| 3,4 5,6 7,8,9 10,11,12 Growth temperature range |
+ ++++ +++ ++ |
+ ++++ +++ ++ |
+ ++++ +++ ++ |
+ ++++ +++ ++ |
| 4 20 37 40,45,50 55 ≥ 60 |
- ++ +++ ++++ +++ - |
- ++ +++ ++++ +++ - |
- ++ +++ ++++ +++ - |
- ++ +++ ++++ +++ - |
| Utilization of avicel | + | + | + | + |
| Utilization of cellobiose | + | + | + | + |
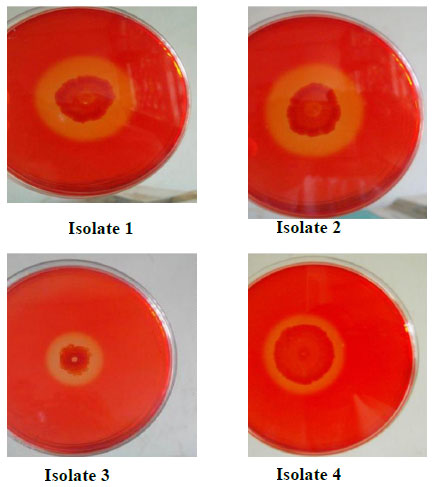
Screening for cellulolytic bacteria by covering the petri dishes with Congo red dye. A zone of clearance surrounding the colonies is indicative of carboxymethylcellulose (CMC) hydrolysis by secreted CMCase.
The cell free supernatant from isolate 3bears CMCase, exoglucanase and β-glucosidase activities.This agrees with the proposed mode of action on cellulosic biomass degradation whereby endo-β-glucanase randomly cleave the cellulose chain to produce cellooligosaccharides, exoglucanase on the other hand act on exposed chain ends to split off cellobiose. Finally, cellobiose is hydrolysed by β-glucosidase to form glucose [2].
Observations that members of the Bacillus genus have a complete cellulase enzyme system have been made by [26]. The cell free supernatant from Bacillus subtilis strains SL9-9, C5-16, S52-2, and KACC10111 were found to bear CMCase and exoglucanase activities only. β-glucosidase activity was however present on the cell debris of the isolates studied [26].

A Phylogenetic tree based on 16S rDNA gene sequences.The four bacterial isolates clustered with Bacillus licheniformis strains suggesting they were likely to be Bacillus licheniformis. Accession numbers KF737352 - KF737355 correspond to the partial 16S rDNA sequences submitted to the Genebank for isolates 1 - 4.
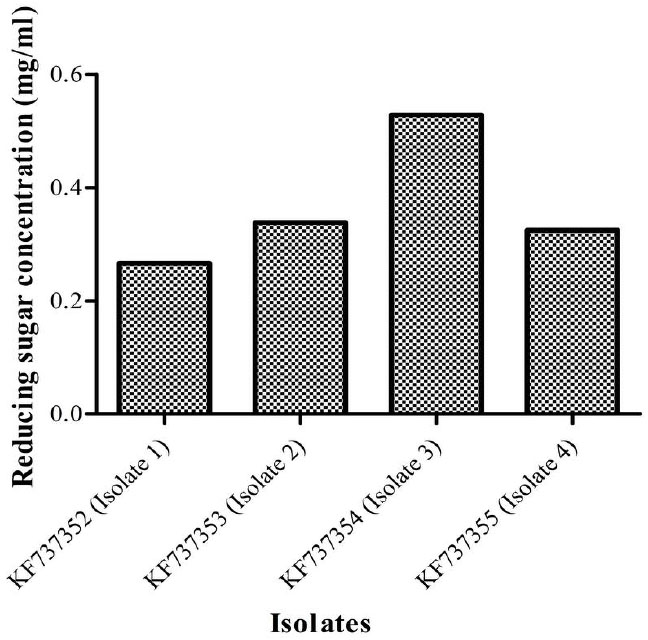
Quantities of reducing sugar (mg/ml) produced by the four isolates. Isolate 3(KF737354) was found to produce the highest concentration of reducing sugars (0.5284 mg/ml) and thus selected for further studies.
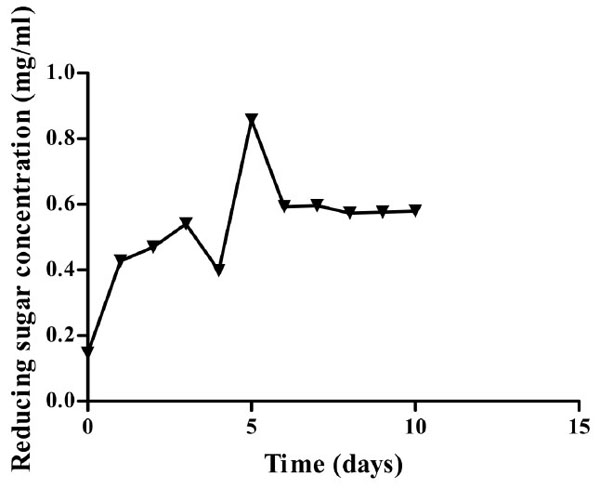
Growth curve of Isolate 3 (KF737354) on minimal media containing CMC as the sole carbon source at 45 oC and pH 6.9. Highest amount of sugar was recorded after five days of incubation.
Optimum Temperature and pH for CMCase Activity
Optimum temperature for Bacillus licheniformis strain Vic was recorded at 60oC with a specific enzyme activity of 0.0698U/mg for the crude enzyme extract (Fig. 5). There was no significant difference in endoglucanase activity at 70oC (P>0). This implies that industrial application of this isolate’s CMCase would yield optimum results at both 60oC and 70oC. Enzyme activity at both 80oC and 90oC was higher than that observed at 30oC. Cellulases of this isolate can thereby be considered to be thermotolerant.
It was noted that optimum temperature for the crude enzyme activity is higher than the optimum temperature for growth of the isolate (Table 1 & Fig. 5). This implies that for industrial applications requiring high temperatures it would be advisable to use the extracellularly excreted enzymes rather than the whole organism.
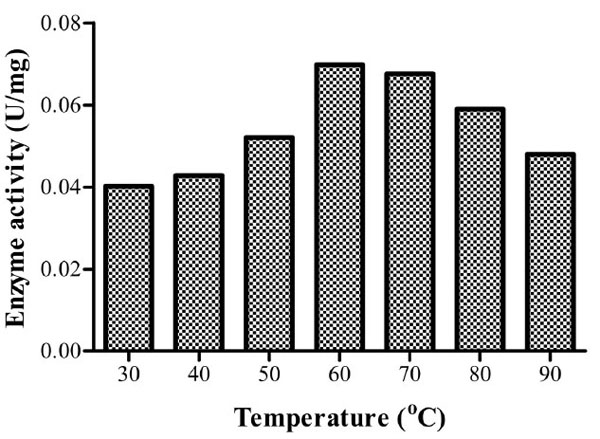
Specific enzyme activities of isolate 3 at different temperatures. Optimum activity was recorded at 60oC.
Several studies have reported the optimum temperature above 50oC for CMCase activity. An optimum temperature of 65oC for Bacillus licheniformis M9 has been reported [10] while Bacillus licheniformis JKF and Bacillus DUSELR13 have higher optima of 70 and 75oC, respectively [16, 14]. Optimum CMCase activities have been recorded at temperatures ≤ 50oC. For instance, 50oC Bacillus sp. [13], 40oC for Bacillus thuringiensis strains [24] and 30oC for Bacillus licheniformis C108 [9].
At the predetermined optimum temperature (60oC), the optimum pH was found to be 5 with a specific enzyme activity of 0.08214U/mg implying that it’s an acidic cellulase (Fig. 6). Acidic cellulases act at a pH range of 3.8-5.8 [27]. Bacillusstrain M-9 [10] was found to have similar optimum pH. A wide range of optimum pH has been noted with reports of values as high as pH 10 for B. licheniformis C108 endoglucanase [9] and pH 4 for B.thuringiensis var. israelensis (BtI) and B.thuringiensis var. thompsoni (BtH) [24].
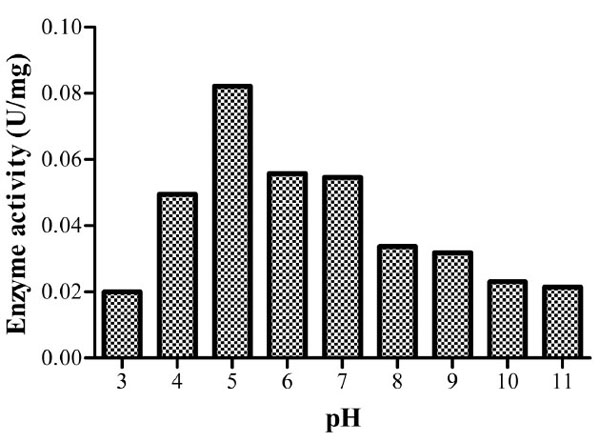
Specific enzyme activities of isolate 3 at different pH values. The optimum activity was recorded at pH 5.
From the time stability study at the predetermined optimum pH and temperature, the isolate’s CMCase activity was found to be stable throughout the study period with 92.2% activity retained after eight hours of incubation (Fig. 7). From the curve, enzyme activity at 6, 7 and 8 hours appears to have reached a platue. Post hoc analysis (Tukey’s test) show that there is no significant difference in the enzyme activity (P>0.05) at this time range. This phenomenon can be attributed to end product inhibition. Cellulase activity can be inhibited by accumulation of cellobiose and glucose [28]. To reduce end product inhibition, supplementation of β- glucosidases during hydrolysis or removal of reducing sugars by ultrafiltration or simultaneous saccharification and fermentation (SSF), can be done [28].
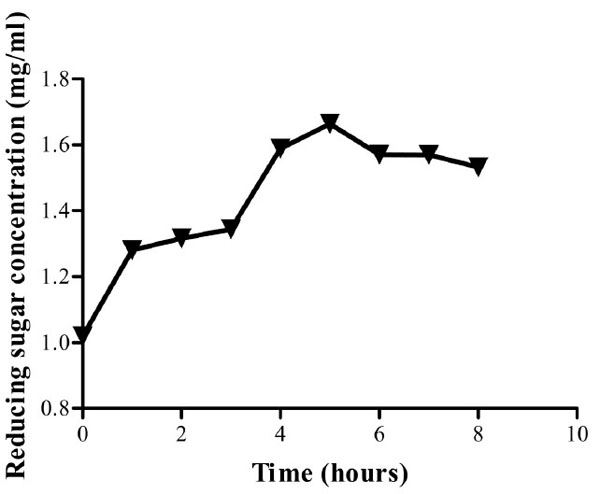
Reducing sugar concentration for enzyme assay up to eight hours. Optimum activity was obtained after five hours. The crude enzyme extract retained 92.2 % of its activity at the eighth hour of incubation at pH and 60oC.
Cellulases from B.licheniformis strain vic can be used to break down cellulose at a high temperature and are stable for relatively long periods of time. This in addition to the extracellular production of the three enzyme components of a cellulase system implies that the isolate can be solely used to produce fermentable products. Bacillus species have the Generally Recognised as Safe (GRAS) status, high growth rate and ability to secrete proteins extracellularly.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
The authors wish to acknowledge International Foundation for Science (IFS) for providing grants for this research and the Postgraduate Laboratory headed by Professor Francis Mulaa in Biochemistry department; University of Nairobi. Additionally, the author wishes to thank the Insect Physiology and Ecology (ICIPE) and WFS national scholarship program- for supporting the researcher with a living stipend.


