All published articles of this journal are available on ScienceDirect.
Production of Ligninolytic Enzymes from Penicillium Sp. and Its Efficiency to Decolourise Textile Dyes
Abstract
Background:
The present study discussed the bio decolourization of synthetic textile dyes using extracellular crude laccase from an Ascomycetes fungus Penicillium sp. Laccase based decolourization is found to be potentially advantageous to bioremediation technologies.
Methods:
In this study, the production of laccase was observed for 7 days of incubation under shaking conditions. Maximum laccase production was secreted by fungal strain on the 6th day of incubation under submerged fermentation. Incubation of fungal mycelium and culture filtrate as crude enzyme obtained from Penicillium sp. with textile dyes - Indigo, Reactive black-5, Acid blue -1 and Vat brown -5 on solid PDA medium and liquid PDA broth showed effective biological dye decolourisation.
Result:
Solid state dye decolourisation had shown 45%, 25%, 50% and 72% colour removal of dyes - Indigo, Reactive black-5, Acid blue -1 and Vat brown -5 whereas maximum decolourization of same dyes of 45%, 20%, 48%, and 75% was obtained in liquid state with crude enzyme within 3h.
Conclusion:
The results had shown the potential dye decolourisation capacity of the Penicillium sp. extracellular crude laccase and pave a way to apply this strain on an industrial scale.
1. INTRODUCTION
Laccase (EC 1.10.3.2, p-diphenol: dioxygen oxidoreductase), a major biocatalyst known for decades was first described in 1883 by Yoshida and he first reported it from the exudates of the Japanese lacquer tree, Rhusvernicifera [1]. They have been found in many taxa including plants, fungi, bacteria, and metazoa [2]. Laccases from fungi have been the focus of intense research due to their possible involvement in the transformation of a wide variety of phenolic compounds including the polymeric lignin and humic substances [3]. Laccases can be secreted extracellular or intracellular. Even though their function is different in the various organisms, all of them can catalyse polymerization or depolymerisation processes [4]. Fungi from Basidiomycota, Ascomycota and Deuteromycota phyla are the sources of fungal laccases, from various ecological niches. They have wide applications in industries such as textile, food, pharmaceuticals and synthetic chemistry as well as an eco-friendly approach with respect to the concept of “green chemistry” [5-7]. Synthetic dyes are used extensively in some industries including textile, food, medicine. [8]. Among these industries, the textile industry generates huge effluent and discharge of this effluent is generating a serious problem [9-17]. Disposal of untreated dyeing effluent without treatment is a serious offence which causes both environmental and health hazards [18]. Even though recalcitrant synthetic dyes are generally removed by various chemical methods, biological removal was regarded as an inexpensive and effective way of treatment [19]. The extracellular ligninolytic enzymes produced by fungi in pure or crude form can decolourise various textile dyes. There is much research related to laccase and dyes degradation by various white rot fungi like Funalia trogii [20], Phanerochaete chrysosporium [21, 22], Trametes versicolor [23], Trametes hirsute [24], and Lentinula edodes [24, 25] but Studies on non-basidiomycete fungi that have the ability in production of lignolytic enzymes and degradation of dyes are scanty. Even though these fungi are also efficient for metabolizing a wide range of compounds, particularly by demethylation and oxidation [26], Aspergillus species [27], Cunninghamella elegans [28], Penicillium geastrivorus [29], P.Ochrochloron [27] Penicillium simplicissimum INCQS 40211 [30] and Penicillium oxalicum SAR-3 [31] are found to be successful for removing textile dyes from liquid media, evaluation of more fungal enzymes in remediation of toxic dye effluents is necessary. In this context, the present work was designed to evaluate laccase activity by Penicillium sp through submerged fermentation and decolourisation capability of obtained enzyme against synthetic industrial dyes (Indigo, Reactive black-5, Acid blue -1 and Vat brown -5). Thus, the results obtained had explained the ability of an Ascomycetes fungal bio decolourisation .
2. MATERIALS AND METHODS
2.1. Fungal Collection Site Description
Soil samples, decayed wood and tree barks with fruiting bodies were collected in and around Tirupati, Chittor dist., A.P, India. The samples collected were stored in sterile bags and stored in cool place for further isolation.
2.2. Isolation of Fungal Strain
The present fungus collected from decayed wood was isolated on Potato Dextrose Agar (PDA) medium by placing a small piece of wood and was incubated at 30 ºC. The fungal colony grown on the plate was microscopically observed with lactophenol cotton blue stain and was recorded at 40X for preliminary morphological characterization. Molecular level identification of the fungus was done by 18s rDNA sequencing and sequence was searched with Blast. The fungus has been assigned the Accession no. and deposited in Genbank.
2.3. Lignolytic Enzymes Production
The isolated fungal strain was screened for lignolytic enzymes production by solid plate method and quantitatively by submerged fermentation. It was done with the inoculation of direct fungal mycelium and spore suspension (2x106 spores/ml) from a 7-day grown PDA slant. Initially, for the qualitative plate assay, a small piece of direct mycelium was placed on PDA plate supplemented with 0.01% guaiacol and incubated at 30ºC for oxidation of guaiacol. Laccase efficiency was calculated by the formation of diameter of brown zone around the fungal colony. Further quantitative estimation of laccase, Lignin peroxidase and Manganese peroxidase activities was done by a 7-day grown fungus (2x106 spores/ml) of spore suspension in liquid PDA broth with 0.01% guaiacol at 150 rpm for 10 days. The culture filtrate was withdrawn at regular intervals after centrifugation at 5000 rpm for 5 min which was served as crude enzyme and was tested for activity of laccase, Lignin peroxidase and Manganese peroxidase enzymes.
2.4. Enzyme Assays
Assays of listed ligninolytic enzymes activities in the cultural filtrate were done by standard protocols. The reaction mixture containing 0.5ml of crude culture filtrate with 10 mM guaiacol in 10% (V/V) acetone containing 100 mM acetate buffer (pH 5.0) was incubated for 5 min and was assayed for Laccase activity. The change in absorbance of the reaction mixture was monitored at 470 nm (ε = 6740 M-1 cm-1) and expressed as Laccase activity [32]. One international unit of activity corresponded to the amount of enzyme that oxidized one micromole of guaiacol per minute. Lignin peroxidase activity was determined within reaction mixture having 0.5 ml culture filtrate, 5 mM H2O2 and 0.25M tartaric acid (pH 2.5) and 2 mM veratryl alcohol. The enzyme activity was expressed in IU where one unit of Lignin Peroxidase corresponded to the amount of enzyme that oxidized one micromole of veratryl alcohol per min [33]. Manganese Peroxidase activity was quantified using a reaction mixture containing crude enzyme 1mM guaiacol, 10mM citrate phosphate buffer (pH 5.5), 1mM MnSO4, and 50 µM H2O2 in a total volume of 1 ml [34]. The reaction was monitored by measuring the increase in absorbance of the reaction product at 465 nm. One unit of Manganese Peroxidase was defined as the amount of enzyme that increased the absorbance at 465 nm by 1.0 per min.
2.5. Dye Decolourisation Potential (Solid and Liquid Studies)
Dye decolourisation potential of the isolated strain was carried out according to the method of Kiiskinen et al. [35]. Indigo, Reactive black-5, Acid blue -1 and Vat brown -5 (each dye at a concentration of 100mg/L) were added individually to 25 ml of 4% potato dextrose agar medium in Petri plates. The pH was adjusted to 5.5 before autoclaving at 121 °C for 15 min. Two millilitres of each dye from stock dyes were added to the media after autoclaving as sterile-filtered water solutions. The positive reaction on the plate was visualized after decolourisation within 3-5 days of incubation at 30 °C and percentage of decolourisation clearing zone was assessed.
The fungal strain was also examined for their ability to decolourise Indigo, Reactive black-5, Acid blue -1 and Vat brown -5 based on the modified method of Eichlerova et al. [36]. The 200 ml Erlenmeyer flask supplemented with above said concentration of dyes were inoculated with working culture filtrate(crude extract). To quantify dye decolourisation by UV visible spectrophotometer at 556nm, 640nm, 600nm, 455nm which are maximum absorbance wavelengths of dyes Indigo, Reactive black-5, Acid blue -1 and Vat brown -5. The samples were prepared in duplicate with 2 ml of each dye and 1ml of crude enzyme and a control was also made without the addition of culture filtrate. Percentage of dye decolourisation was calculated using the formula as shown below:
Percentage of dye decolourisation (%) = R % = [(Initial absorbance - final absorbance) / (initial absorbance)] x 100
Where the initial absorbance of the untreated dye and the final absorbance indicated absorbance of dye after treatment with enzyme source.
3. RESULTS AND DISCUSSION
3.1. Morphology, Growth Characters of the Fungus
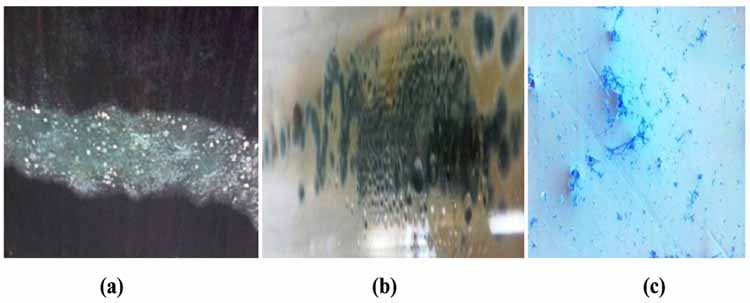
After 5 days of incubation on PDA medium, the fungal culture was grown as greyish green colonies and designated as ‘W’ (Figs. 1a and b). The microscopic examination revealed the development of septate mycelium with many branched conidiospores (2-3μm). Based on colony morphology and microscopic features, the fungus was identified as Penicillium (Fig. 1c). Molecular characterisation was done by using ITS 4 and ITS 6 primers and was found that the sequence had a similarity with GU565119.1, GU565118.1 etc and was belonging to Ascomycetes family, thus, the strain ‘W’ based on morphological and phylogenetic analysis was identified as Penicillium sp. and was deposited in GenBank under accession no: KY930467.1 (Fig. 2).

3.2. Ligninolytic Efficiency of Penicillium Sp.
To check for the ability of the present strain for the production of laccase, the strain was grown in PDA medium with guaiacol both on slants and plates. The grown fungal colonies on slant had shown oxidation of guaiacol on the first day itself (Fig. 3) and produced reddish brown zone around and beneath the colony on PDA slant and plate due to oxidation of guaiacol. The positively screened plate photo was not placed and shown here due to a technical problem. The ligninolytic efficiency was further quantified in the same medium under shaking conditions with an rpm of 180 for the production of laccase, lignin peroxidase and manganese peroxidase enzymes, respectively. Production of laccase was maximum after 5 days of incubation and reached maximum on the 6th day. Later, the enzyme production was decreased from 7th day and was shown in Table 1. Lignin peroxidase and manganese peroxidase enzymes production by this strain were observed negative.

| Incubation Days | Laccase Activity(U/Ml) |
|---|---|
| 1 | 0.1 |
| 2 | 0.6 |
| 3 | 7.1 |
| 4 | 12.6 |
| 5 | 10.4 |
| 6 | 8.3 |
| 7 | 6.1 |
| 8 | 5.2 |
| 9 | 3.1 |
| 10 | 2.4 |
The factors influencing the LiP or MnP or laccase production may depend on different conditions like strain or culture conditions. Negative LiP or MnP or laccase production in some strains may suggest that these fungi either produce no significant levels of these enzymes or their production requires different conditions. These type of results were true and studied for several strains of Trametes versicolor, Bjerkander aadusta which are known as LiP producers but do not always produce lignin peroxidase depending on strain or culture conditions [37, 38]. In addition, previous results indicated the production of laccase reaches its maximum value with a delay of twenty-sixth day of incubation with a specific activity of 7.18 U/mg protein in a relatively gradual manner and then falls by Penicillium martensii NRC 345 [39]. Though the degradation of lignocellulosics by ligninolytic enzymes of Penicillium sps. is different from that of the white-rot fungi [40], some reports had described the detection of the laccase, MnP and LiP enzymes by P. pinophilum TERIDB1 and A. aisen TERIDB6 was described by Pant et al. [41]. A similar type of work and secretion of ligninolytic enzymes was observed by Sack and Gunther, 1993 [42]. Recently Yang et al. reported the production of LiP by Penicillium decumbens [29]. In contrast, Zeng et al stated that the production of ligninolytic enzymes by most white-rot fungi was found to be a ‘secondary metabolic’ event [43]. Concerning the delay of optimum laccase formation which reached the 26thday of incubation and reported that Botrytis cinerea produced appreciable levels of laccase (9.8 U/ml) in a brief period (5-7 days) and they stated that, some fungal species required a longer production time (12-30 days) [44].
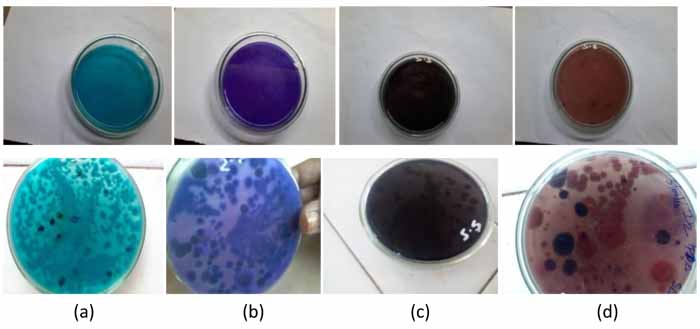
Crude extracellular culture filtrate of present strain Penicillium sp. was tested for the dye decolorizing ability on agar plate amended with four dyes used in this study at a concentration of 100 mg/L. Penicillium sp. was active to various levels in decolorizing the four dyes tested and started decolourization of Indigo and Vat brown -5 on the first day whereas Reactive black-5 and Acid blue -1 after 48hrs and with a range of 25-70% of decolourisation in 3 days and it does not proceed further. During decolourisation removal of dye from the plate was also may be due to adsorption of dye to the mycelium as colony growth was observed on plates. To confirm the dye decoloration was due to minimal adsorption and maximum by enzymatic mechanism, liquid decoloration experiments were carried out.
The crude extract after 7 days from fermented broth was evaluated for enzymatic decolourisation against Indigo, Reactive black-5, Acid blue -1 and Vat brown -5. About 75% decolourisation was observed within 3 hours with Vat brown -5 and 48% in case of Acid blue -1 whereas 45% and 20% in Indigo and Reactive black-5, respectively without any mediators (Figs. 4 and 5, Table 2).
| Dyes | λ Max | % of Dye Decolourisation | |
|---|---|---|---|
| Solid Decolourisation | Liquid Decolourisation | ||
| Indigo | 455 | 45% | 45% |
| Reactive black-5 | 600 | 25% | 20% |
| Acid blue -1 | 640 | 52% | 48% |
| Vat brown -5 | 556 | 70% | 75% |

4. DISCUSSION
Biotechnological methods like isolation of fungi from diverse environments and adopting their enzymes in the degradation of xenobiotics and various dyes are applying globally. The key enzymes that play a major role in bioremediation of environmental pollutants are lignolytic enzymes like manganese peroxidase, lignin peroxidise and laccase. In view of this importance, the present study was undertaken in orde to check the potentiality of the organism in secretion of major extracellular enzymes like laccase, manganese peroxidise and lignin peroxidise and their application in dye decolourisation. Screening different organisms and their enzymes for dye decolorization are beneficial as it may result in more efficient dye decolorizers than the commonly studied organisms. In this context, Penicillium sp. isolated from wood in this study was also found to be a laccase producer and production was increased in short incubation period. Results reported by Artexga et al. and Levin et al. that similar rmycelial results when performing dye degradation experiments using white rot fungi [45, 46]. It is to note in their results that the fungi Peniophora sp.hpF-04 and Phellinus sp.hpF-17 being predominant laccase producers and decolourisation are due to laccase production as well as media components. Several other authors had also discussed the role of enzymes in the decolourisation activity of ligninolytic fungi [47-49]. Number of researchers had worked on fungal decolourization studies to know the main and effective fungal enzymes. Various mechanisms have explained the role of lignin degrading enzymatic system of white-rot fungi and unknown enzymes in fungal dye decolourization. Among the ligninolytic enzymes, laccase is highly studied for its role in decolourisation and detoxification of various industrial and textile dyes. It was also reported that laccase is solely responsible for the decolourisation and degradation of dye [48, 50, 51]. Intracellular enzymes like vanillyl-alcohol oxidase and catalase-peroxidase of P. simplicissimum degraded a variety of compounds. The decolourisation of different dyes by many organisms and particularly various sps of Penicillium are discussed below. About 70% Reactive Brilliant Red X-3B was decolourised by a new strain Penicillium sps QQ [52]. Decolorization of direct blue by Aspergillus flavus and Penicillium canescens was after 6 days incubation in findings of Hefnawy et al. Husseiny [53] who found that the maximum degradation activity of Penicillium spp of reactive and direct dye was at 35 ºC. The findings of Muthezhilan observed that Penicillium citrinum decolorized maximum number of dyes like methylene blue, gentian violet, crystal violet, cotton blue, Sudan black, malachite green, methyl red and corbol fushion to a great extent whereas Penicillium oxalicum expressed very low amount of color reduction [54]. Intracellular enzymes like vanillyl-alcohol oxidase and catalase-peroxidase of P. simplicissimum degraded a variety of compounds [55, 56]. The capacity of decolorization and detoxification of the textile dyes Reactive Red 198 (RR198), Reactive Blue 214 (RB214), Reactive Blue 21 (RB21) and the mixture of the three dyes (MXD) by Penicillium simplicissimum INCQS 40211. Aspergillus flavus and Alternaria solani, the decolorization and degradation of a triphenylmethane dye in 6 days. The study of Zheng et al. observed that Penicillium sp. removed and degraded the dyes Poly R-478 and Poly S-119 [57]. Penicillium oxalicum SAR-3 isolated and screened from dye contaminated soils decolourised and degraded Acid Red 183 (AR 183), Direct Blue 15 (DB 15) and Direct Red 75 (DR 75) at 100 mg L(-1) concentration with its manganese peroxidise enzyme [31]. High decolorization or degradation dyes also takes place either by laccase alone or with mediators [58, 59] and reduces toxicity also [58, 60]. The enzyme producing activity of White rot fungi makes them effective decolorizers and biode-graders [61-66]. Chung and Aust had reported that with laccase-mediator, there was high biodegradation than laccase alone [67]. Mediators such as phenolic compounds, ABTS, HBT, and 3HAA could enhance dye degradation [68]. Laccase of Cyathus bulleri decolorized and detoxified 80% the mixture of reactive and acidic dyes in presence of natural and synthetic mediators [69]. Complex mixtures of dyes were decolorized with an enzyme in the presence of HOBT [70]. Studies of Machado and Husain, 2007 obtained maximum decoloration in a complex mixture of dyes with the enzyme and 0.06 mM redox mediator 1hydroxybenzotriazole/violuric acid [71]. Whereas treatment of anthraquinone dyes with purified enzyme along with mediator degraded azo dyes [72]. Momordica charantia immobilised peroxidase was found to be effective in decolourization textile waste Water [73]. Tricho santhesdioica peroxidase- concanavalin A (PGP-Con A) complex, entrapped into calcium alginate pectin gel decolourised a mixture of two dyes [74]. Some other reports e.g. Rodriguez et al., 2005, Wong and Yu, Gochev and Krastanov, Ismat Bibi are also explained the role of mediator along with laccase in dye decolourisation [75-78]. To remove color from dye effluents purified and/or immobilized enzym is used which is not economical. But the use of effective crude enzyme is economically viable and the present study explained very easy production and preparation of crude enzyme from this fungus to decolorize synthetic dyes without the role of mediators.
CONCLUSION
The Ascomycetes fungus Penicillium sp. had produced laccase under submerged fermentation. Four dyes were bio catalytically decolorized by extracellular laccase secreted by the present strain in solid and liquid cultivation. It was confirmed that the decolourisation was correlated with laccase and that Penicillium sp. was capable of decolorizing a range of industrially important textile dyes. The difference in decolourisation rates was according to dye types and due to various structures of the dyes. In conclusion, fungal decolourisation appears to be a promising alternative to replace currently available chemical treatment processes.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this review.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
First Author, grantee of UGC-PDF is very grateful to the UGC in India for financial support and also thankful to the Sri Padmavathi mahila University authorities for smooth conduct of research work.


