RESEARCH ARTICLE
Isolation and Characterization of Polyethylene and Polyethylene Terephthalate-degrading Bacteria from Jakarta Bay, Indonesia
Aqil Azizi1, *, Sirin Fairus1, Deffi Ayu Puspito Sari1
Article Information
Identifiers and Pagination:
Year: 2024Volume: 18
E-location ID: e18740707280343
Publisher ID: e18740707280343
DOI: 10.2174/0118740707280343231208102253
Article History:
Received Date: 05/09/2023Revision Received Date: 29/10/2023
Acceptance Date: 21/11/2023
Electronic publication date: 06/02/2024
Collection year: 2024

open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction
Plastic is resistant to natural breakdown because of its intricate structure, which features long and repeated molecular chains. As a result, a variety of plastic waste, mostly made of polyethylene (PE) and polyethylene terephthalate (PET), accumulates in Jakarta Bay. The use of microorganisms to degrade plastic trash has emerged as a highly promising bioremediation strategy.
Methods
The goal of this research is to find microorganisms capable of digesting plastic in the samples of seawater and sediment obtained from Muara Angke Jakarta Bay. The bacteria were grown on Zobell Marine Agar (ZMA) that had been treated with 2% polyethylene glycol (PEG). The bacteria were then selected based on their capacity to degrade PE and PET microplastics in a liquid medium. The ability to degrade was determined by measuring the optical density (OD) at 600 nm and the decrease in plastic dry weight after a 14-day incubation period.
Results
Seven bacterial isolates capable of decomposing PE and PET were found during the isolation and screening methods. The WJ1 outperformed other isolates in the degradation of PE and PET, with degradation rates of 4.5% and 6.5%, respectively.
Conclusion
According to 16S rRNA analysis, five bacterial species have been identified as playing a part in the process of plastic degradation: Vibrio alginolyticus, Pseudoalteromonas caenipelagi, Microbulbifer pacificus, Pseudomonas marincola, and Bacillus subtilis. The ability of these bacteria to biodegrade plastics represents an opportunity to effectively remove persistent pollutants from the environment.
1. INTRODUCTION
There is a significant prevalence of plastic pollution in the Earth's oceans [1, 2]. According to available reports, an estimated annual influx of plastic debris into the ocean ranges from 4.8 to 12.7 million tons [3]. The issue of plastics in the marine environment is becoming increasingly concerning due to their long-lasting presence and the negative impact they have on the oceans, wildlife, and potentially humans. Furthermore, following exposure to weathering, mechanical abrasion, and ultraviolet radiation, the significant plastic material may undergo fragmentation, resulting in particles smaller than 5 mm in diameter. These particles are commonly referred to as microplastics [4].
The vast majority (46.5% by weight) of plastic pollution consists of two types of plastic: polyethylene (PE) and polyethylene terephthalate (PET) [5]. Because of the non-hydrolyzable covalent links in its carbon-carbon backbone [6], PE is very resistant to mechanisms that break it down. PET, a semi-aromatic thermoplastic co-polymer resin, is a member of the polyester family [7]. The primary chain's distinctive characteristics are derived from its composition of heteroatoms and aromatic groups. In many ways, PE and PET are interchangeable. Some examples are a high molecular weight, a low density, a lengthy hydrocarbon chain, and a high tensile strength. They are non-biodegradable, resistant to physical and chemical breakdown, and have a low permeability to gases [8-10].
One possible method for converting non-biodegradable waste petro-plastics into recyclable monomers is the degradation of plastics by microbial and enzymatic activities. Plastics can also be mineralized through these processes, yielding carbon dioxide, water, and new biomass as byproducts. This modification can result in the creation of significant bioproducts, as previous research [11-13] has demonstrated. Microorganisms secrete extracellular enzymes to break down plastic throughout the biodegradation process. Once attached to the plastic, these enzymes trigger hydrolysis and the production of shorter polymer intermediates on the plastic's surface. Microorganisms use these intermediaries as a source of carbon, which ultimately results in the production of carbon dioxide. Despite the synthetic nature of plastics, numerous microbes that are able to metabolize them have been discovered in recent years [14].
Recent research that identified and measured the amount of microplastics in the coastal water and sediment of Jakarta Bay showed that there are a lot more microplastics in Jakarta Bay than in other water areas in Indonesia. The two most common types of microplastic polymers were PE and PET [15]. Based on the amplicon sequencing of the 16S rRNA gene, the data from the same area also show a wide range of sulfur-oxidizing bacteria colonializing in two types of microplastics. Sulfurovum sp., a chemolithoautotroph bacterium, colonizes and dominates the entire bacterial population [16]. It has been suggested that a subset of the microbial population that colonizes plastics may be able to degrade plastics and use them as a carbon matrix because some marine bacteria are capable of breaking down hydrocarbons that have a chemical composition similar to plastics [17]. In this study, we conducted a screening and isolation process to identify bacteria capable of degrading microplastics. The samples used for this investigation were collected from both seawater and sediment in the Muara Angke Jakarta Bay, which is known to be contaminated by plastic waste.
2. MATERIALS AND METHODS
2.1. Sampling
Samples of seawater and sediment have been taken from the coastal region of Muara Angke, located in Jakarta Bay (Fig. 1a, b). A total of 100 g of sediment samples were collected with the Eijmen grab sampler from a depth of 30 cm below the water's surface, utilizing purposive random sampling. The sediment is placed in a plastic ziplock bag and labeled with the location, time, and date of collection. A 20 L water sample was also obtained and filtered many times with a plankton net to get a 1 L seawater sample. All samples were placed in a cold box at 4oC for further analysis in the laboratory.
2.2. Isolation and Characterization of Bacteria
Both seawater and sediment were subjected to the dilution series. From dilutions 10-6, 10-7, and 10-8 of sediment and seawater samples, 100 μL were taken and spread in a petri dish containing Zobell Marine Agar (ZMA), to which 2% polyethylene glycol (PEG) had been added to investigate the ability of isolates to grow in a plastic-containing environment. The dish was then incubated for 48 hours at 37oC. A series of successive inoculations were conducted on ZMA media supplemented with 2% PEG until a culture of high purity was achieved. The examination of colonies and cells involved the observation of their morphology, specifically focusing on characteristics, such as color, shape, edge, and elevation.
2.3. Biodegradation Test
The bacteria were then re-inoculated as pure cultures on fresh ZMA liquid media and cultured at room temperature for 14 days in a 150-rpm shaker. For the microplastic degradation test, 10% of the culture was inoculated into a 250 ml Erlenmeyer flask containing 100 ml of ZMA liquid medium and 0.5 g of 2.4 mm-diameter PE and PET microplastics. Bacterial growth was assessed using a spectrophotometer to measure the optical density at 600 nm. Microplastics were cleaned with 70% ethanol before being dried. An analytical balance was used to weigh the microplastics. The formula [18] is used to calculate the percentage breakdown of plastic waste samples by bacterial culture:
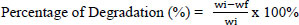 |
wi = Dry weight before degradation (g); wf = Dry weight after degradation (g)
2.4. DNA Sequence Analysis
The GeneAll DNA exgene CV cell was utilized for DNA extraction. 16S rRNA gene amplification was carried out using the primer pairs 27F (5'-AGAGTTTGATCMTGGTCCAG-3') and 1492R (5'-TACGGHTACCTTGTTACGACTT-3'). Amplification was carried out with a PCR reaction composition consisting of 3 µl DNA template, 12.5 µl DreamTaqTM Green PCR Master Mix (2x) (Thermo Scientific, Lithuania), 1 µl forward primer, 1 µl reverse primer, and 7.5 µl nuclease-free water, for a total of 25 µl. The PCR amplification process was carried out in the Arctic Thermal Cycler (Thermoscientific) with pre-denaturation conditions at 96 ˚C for 5 minutes, denaturation at 96 ˚C for 30 seconds, attachment at 55 ˚C for 30 seconds, elongation process at 72 ˚C for 60 seconds, and final elongation at 72 ˚C for 7 minutes. The PCR process lasts for 30 cycles. The amplification results were then purified and sequenced using the services of Genetika Science Indonesia. The sequences were aligned with representative reference sequences (version 1.83) using CLUSTAL X. Phylogenetic trees were created with the MEGA program (version 69.05). Neighbor-joining tree topology was assessed using a 1,000-repeat bootstrap technique [19].
3. RESULTS AND DISCUSSION
3.1. Isolation of Bacteria Capable of Degrading PE and PET
Based on the results of repeated culture and purification, 10 bacterial isolates with the capacity to break down microplastics were found in seawater and sediment samples. Numerous factors, including the presence of organic matter and environmental conditions like salinity, pH, temperature, depth, and current speed, might contribute to variations in the number of bacterial isolates in a given location. The plastic monomer PEG is added to the isolation medium to determine the ability of microbes to degrade plastic. It can be presumed that bacteria that grow in a medium containing PEG have the ability to degrade microplastics.
Bacterial colony observations revealed morphological differences across bacterial isolates (Table 1). The variation in bacterial colony growth implies that the bacterial isolates were most likely from different bacteria. Based on the observations, two bacterial isolates were white, four bacterial isolates were clear white, two bacterial isolates were yellowish white, one bacterial isolate was milky white, and one bacterial isolate was creamy. The bulk of bacterial colony isolates were circular, with only two exceptions being irregular in shape. Microscopic cell morphological studies were carried out in addition to macroscopic observations to identify cell shape and differentiate gram-positive and gram-negative bacteria. Gram-negative and gram-positive staining is a method of categorizing bacteria based on the peptidoglycan content of their cell walls. The peptidoglycan layer in gram-positive bacteria is thicker, whereas the peptidoglycan layer in gram-negative bacteria is thinner. Gram-positive bacteria retain a rich purple color following alcohol washing, whereas gram-negative bacteria do not [20, 21]. All of the bacterial cell isolates in this study were basil-shaped and gram-negative.
| Isolate | Colony Morphology | Cell Morphology | |||||
| Size | Pigmentation | Shape | Edge | Elevation | Shape | Gram | |
| SJ1 | Medium | Yellowish white | Circular | Flat | Droplets | Bacil | - |
| SJ2 | Medium | Clear white | irregular | Curved | Flat | Bacil | - |
| SJ3 | Medium | Clear white | Circular | Wavy | Flat | Bacil | - |
| SJ4 | Small | Creamy | Circular | Flat | Flat | Bacil | - |
| SJ5 | Medium | Clear white | Circular | Curved | Flat | Bacil | - |
| SJ6 | Medium | Clear white | irregular | Wavy | Flat | Bacil | - |
| SJ7 | Medium | White | Circular | Curved | Convex | Bacil | - |
| WJ1 | Medium | Milky white | Circular | Curved | Flat | Bacil | - |
| WJ2 | Large | Yellowish white | Circular | Wavy | Convex | Bacil | - |
| WJ3 | Medium | White | Circular | Wavy | Droplets | Bacil | - |
3.2. Selection of Bacteria with the Capability of Degrading PE and PET
Only seven of the ten bacterial isolates isolated on ZMA media containing 2% PEG exhibited optimal growth, namely SJ1, SJ3, SJ4, SJ7, WJ1, WJ2, and WJ3. The seven bacterial isolates were tested for their ability to degrade microplastics. OD was used to measure the growth of seven bacterial isolates on media containing PE and PET microplastics for 14 days, as shown in Fig. (2a, b). On day 6 in both PE and PET-containing media, 2 isolates (WJI and SJ3) exhibited maximal growth. Meanwhile, isolate WJ2 reached its maximum growth on both forms of media on day 8. Initial inoculation did not result in optimum growth for both isolates SJ1 and WJ3. The growth of bacteria in media containing plastic polymers and several other nitrogen elements indicates that these microbes can utilize polymers to meet their carbon requirements in metabolic processes by releasing catalytic enzymes that can damage polymer structure [20].
3.3. Microplastic Biodegradation
In this study, the ability of bacterial isolates to degrade microplastics was enhanced by the fact that the bacteria isolated originated from areas with plastic waste, including microplastics, and that 2% PEG was added to the isolation medium. The biodegradation test was done by finding the percentage difference between the starting and ending weights of the PE and PET microplastics in the medium with bacteria (Table 2).
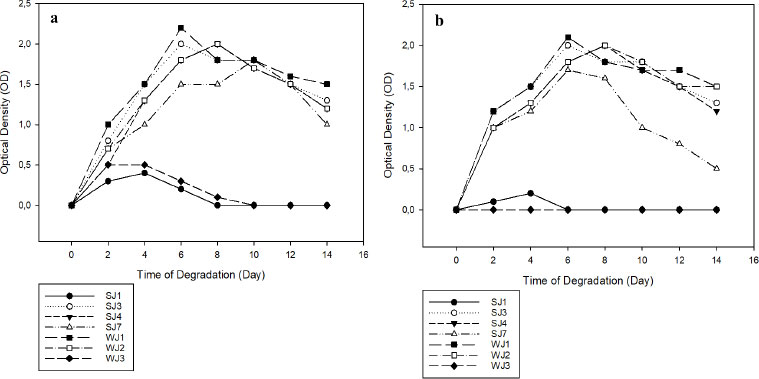 |
Fig. (2). Graph of OD values. Inoculation with PE (a); Inoculation with PET (b). |
| Isolate | PE | PET | ||||||
| Initial dry weight (g) | Final dry weight (g) | Subtraction (g) |
% Degradation |
Initial Dry Weight (g) | Final Dry Weight (g) | Subtraction (g) |
% Degradation |
|
| SJ1 | 0.522 | 0.52 | 0.002 | 0.4 | 0.502 | 0.5 | 0.002 | 0.4 |
| SJ3 | 0.537 | 0.517 | 0.02 | 3.7 | 0.515 | 0.49 | 0.025 | 4.9 |
| SJ4 | 0.525 | 0.512 | 0.013 | 2.5 | 0.527 | 0.505 | 0.022 | 4.2 |
| SJ7 | 0.542 | 0.527 | 0.015 | 2.8 | 0.535 | 0.512 | 0.023 | 4.3 |
| WJ1 | 0.515 | 0.492 | 0.023 | 4.5 | 0.528 | 0.495 | 0.033 | 6.3 |
| WJ2 | 0.509 | 0.498 | 0.011 | 2.2 | 0.548 | 0.518 | 0.03 | 5.5 |
| WJ3 | 0.553 | 0.55 | 0.003 | 0.3 | 0.513 | 0.513 | 0 | 0 |
Based on the tests conducted over a period of 14 days, it was observed that a number of microplastic samples exhibited a decrease in weight relative to their starting weight. There are five bacterial isolates that possess the capability to participate in PE and PET microplastic degradation processes. These isolates are identified as SJ3, SJ4, SJ7, WJ1, and WJ2. The respective percentages of PE degradation for these isolates are 3.7% (SJ3), 2.5% (SJ4), 2.8% (SJ7), 4.5% (WJ1), and 2.2% (WJ2). Similarly, the percentages of PET degradation for these isolates are 4.9% (SJ3), 4.2% (SJ4), 4.3% (SJ7), 6.3% (WJ1), and 5.5% (WJ2). In contrast, the degradation results of isolates SJ1 and WJ3 were found to be insignificant since there was no observed drop in their initial weight, as seen in Table 2. Based on the provided data, it is noticeable that WJ1 demonstrates the highest degree of degradative activity among the isolates.
The substrate is one of many elements that can affect the degradation process. The substrate refers to the composition and size of the constituent compounds present. If the size of the substrate is reduced and the complexity of the constituent compounds is decreased, the rate of degradation will be accelerated. Additionally, factors like a polymer's chemical composition, molecular weight, and degree of crystallinity all affect how quickly it biodegrades. Polymers are characterized by their molecular structure, which consists of both crystalline regions and amorphous regions. The presence of these irregular groups in the polymer structure imparts flexibility to the material. When the degree of crystallinity in a polymer, such as PE, is significantly elevated (95%), it results in a rigid material that exhibits limited resistance to impact.
In contrast, plastic derived from PET exhibits a notable degree of crystallinity, ranging from 30% to 50%. This characteristic significantly impedes microbial degradation, resulting in an estimated degradation period of approximately 50 years within the natural environment. However, when PET plastic is disposed of in marine ecosystems, its degradation time extends to hundreds of years. The decrease in temperature and the limited supply of oxygen have been identified as contributing factors [14]. The study of bacteria breaking down PE is much less advanced than that of breaking down PET, and breaking down with a single strain or enzyme is still not good enough for industrial use [22].
The findings of this study indicate that PE microplastic substrates exhibit a lower degradation value in comparison to PET microplastic degradation values. This disparity is believed to be the underlying cause for the limited degradation activity observed in some bacterial isolates when exposed to PE microplastics. During the initial stages of the degradation process, the polymer undergoes conversion into monomers, which then undergo mineralization. Due to their size, the majority of polymers are unable to traverse cell membranes, necessitating their mineralization into smaller monomers prior to being assimilated and metabolized by microbial cells. The mineralization process typically leads to alterations in the characteristics of the polymer, including the breaking, transformation, or synthesis of new chemical bonds within the polymer structure. The process of depolymerization involves the breakdown of intricate plastic polymers into smaller units, such as monomers, dimers, and oligomers. This breakdown is facilitated by depolymerase enzymes, which can be found both extracellularly and intracellularly. Consequently, these molecules possess the ability to be readily transported into bacterial cells, serving as both an energy and carbon source [23].
3.4. Identification of Bacteria Capable of Degrading PE and PET
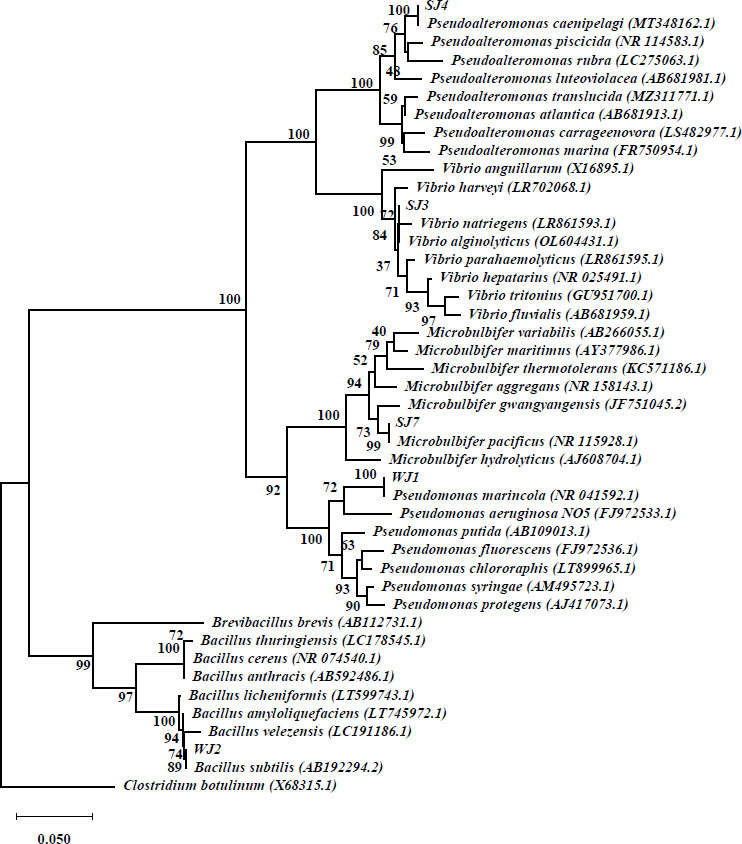
Based on the results of the 16S rRNA gene identification, it was observed that three isolates exhibited a similarity level of over 99%. As per the guidelines proposed by [24], the criteria utilized for species identification entail assessing sequence similarity, with a minimum threshold of 99% and an optimal threshold of 99.5%. In order to achieve identification at the genus level, percentages ranging from 97% to 98% are commonly employed. One isolate was identified to share the same genus as the reference species, perhaps indicating the presence of a novel species [25]. In addition, it is worth noting that one isolate exhibited dissimilarity at both the species and genus levels, as its similarity score fell below 97%. Consequently, this particular bacterial isolate may potentially represent a novel species, necessitating further investigation for proper identification (Table 3). The findings of the phylogenetic analysis, which utilized the Neigbor Joining technique and relied on 16S rRNA sequences, are presented in Fig. (3).
Table 3. Results of nucleotide base alignment analysis of the 16S rRNA gene with the nucleotide database in GenBank.
| Isolate | Accession Number | Scientific Species Name | Quary Cover (%) | Similarity Rate (%) |
| SJ3 | MT325883.1 | Vibrio alginolyticus | 98 | 99.10 |
| SJ4 | MT348162.1 | Pseudoalteromonas caenipelagi | 99 | 96.33 |
| SJ7 | NR_115928 | Microbulbifer pacificus | 100 | 99,39 |
| WJ1 | KY940349.1 | Pseudomonas marincola | 99 | 99,01 |
| WJ2 | ON514287.1 | Bacillus subtilis | 99 | 98.25 |
One of the bacteria that degrade PE and PET in this study is classified under the genus of Vibrio. Vibrio is a type of bacteria that has a rod-shaped shape, grows in mesophilic conditions, needs chemoorganotrophic food, and can also use anaerobic metabolism. Its cell walls are Gram-negative. Vibrios are commonly observed in aqueous habitats, particularly marine environments, and are recognized for their ability to thrive either independently or as symbiotic organisms with aquatic animals [26]. In this study, Vibrio sp. was isolated and subsequently identified as V. alginolyticus. In a study conducted by [27], it was documented that a consortium of Vibrio sp., specifically V. alginolyticus and V. parahemolyticus, was identified as being capable of degrading low linear density polyethylene (LLDPE) combined with 30% polyvinyl alcohol [28-34].
The strains of Pseudoalteromonas found in this study are 96% similar to Pseudoalteromonas caenipelagi, which suggests that they probably evolved separately, mostly at the genus level. Therefore, additional research is necessary. The Pseudoalteromonas genus comprises bacteria that are gram-negative, heterotrophic, aerobic, and motile. It is capable of synthesizing extracellular enzymes, antibiotics, and polysaccharides as well as exhibits the ability to thrive in low-temperature conditions and demonstrates a notable tolerance to high salinity levels [34]. In a study conducted by [35], it was shown that the presence of Pseudoalteromonas was detected in samples of PE plastic through the utilization of a metabarcoding technique. Pseudoalteromonas species exhibit a broad distribution throughout many maritime habitats, including sediments, coastal regions, and estuaries. According to prior studies [36], Pseudoalteromonas caenipelagi has been documented in coastal marine environments.
Microbulbifers are frequently found in environments with high salinity, such as mangrove soils, coastal sediments, and marine habitats [28-30]. M. Pacificus was found in maritime habitats throughout Korea in a previous study [31]. Similarly, another study [32] discovered M. salipaludis in coastal environments in Saudi Arabia. The capacity of the Microbulbifer genus to degrade LLDPE-type PE has been demonstrated through the observation of morphological alterations on the surface of the PE material [33].
Pseudomonas is a genus of bacteria that can be found in a variety of environmental niches, including soil, water, and plants [37, 38]. P. marincola has been detected in numerous marine habitats, according to a study undertaken by [39]. The previous study [36] indicates that some species of Pseudomonas exhibit biodegradation activity towards low-density polyethylene (LDPE), resulting in a degradation rate of up to 20% over a period of 120 days. Meanwhile, P. alcaligenes, a bacterium isolated from plastic-polluted soil, has been found to break down PE by 20% over a 30-day period [40]. Pseudomonas possess the capacity to synthesize serine hydrolase, esterase, and lipase enzymes, which have the capability to break down plastic materials. The enzymatic degradation of plastic trash is most effective when there are no environmental inhibitors that impede enzyme activity (20). Pseudomonas demonstrate an active capacity to adhere to and establish a biofilm on the surface of plastic trash as part of the biodegradation process.
Bacteria belonging to the genus Bacillus, specifically Brevibacillus spp., have the ability to degrade PE in order to acquire carbon sources. Brevibacillus species have the ability to form biofilms on the surface of plastic materials. The process of plastic deterioration is estimated to occur over a period of three weeks, leading to a decrease in the dry weight of the plastic by 37.5% [41]. A study [42] investigated the degradation of polypropylene (PP) using Bacillus sp. and Rhodococcus sp. The results indicated that the strain of Bacillus sp. obtained a weight loss of 6.4% over a period of 40 days, while the strain of Rhodococcus sp. produced a weight loss of 4%.
The predominant bacteria species reported in the previous study on the seawater and sediments of Muara Angke, Jakarta Bay, were not the same as the isolates obtained in this study. According to previous studies, Sulfurovum lithotrophicum is the most prevalent microbe colonized in PE and PET microplastics in Muara Angke, Jakarta Bay, an area extensively polluted by plastic and hydrocarbon pollutants such as oil, diesel, and gasoline [17]. Meanwhile, no elemental sulfur or thiosulfate was added to the culture medium used in this study to serve as the sole source of electrons for the sulfur-oxidizing bacteria. This explains why sulfur-oxidizing bacteria do not proliferate.
CONCLUSION
A screening and isolation process was conducted to identify bacteria capable of degrading microplastics in seawater and sediment from Muara Angke Jakarta Bay. Over 14 days, microplastic samples showed a decrease in weight. Five bacterial isolates, SJ3, SJ4, SJ7, WJ1, and WJ2, were found to participate in PE and PET microplastic degradation activities. These isolates showed varying percentages of PE and PET degradation. SJ1 and WJ3 showed no significant degradation, while WJ1 showed the highest degrading activity (4.5% and 6.3% for PE and PET, respectively). Five bacterial species, Vibrio alginolyticus, Pseudoalteromonas caenipelagi, Microbulbifer pacificus, Pseudomonas marincola, and Bacillus subtilis, are identified in plastic degradation. Future studies should further improve polymer degradation efficiency and explore various pretreatment strategies. Furthermore, specific mechanisms of microorganisms involved in plastic biodegradation also need to be thoroughly investigated.
LIST OF ABBREVIATIONS
| PE | = Polyethylene |
| PET | = Polyethylene Terephthalate |
| ZMA | = Zobell Marine Agar |
| OD | = Optical Density |
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data and supportive information are available within the article.
FUNDING
This research was funded by the Grant of the Directorate General of Higher Education (DIKTI) Republic of Indonesia 2023. It was also supported in part by Kurita Asia Research Grand (22Pid114) provided by Kurita Water and Environment Foundation.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.